The Download Genome Reference dialog allows you to download and import template sequences directly from the National Center for Biotechnology Information (NCBI). This dialog is accessed by either:
•Pressing the Download button in the Set Up Preprocessing screen of an RNA-Seq, ChIP-Seq, miRNA or CNV project.
•Choosing Data > Download Genomic Template from the main menu.
In the dialog, you first need to choose whether to download by accession number, or to download a whole genome by organism name.
Downloading by accession number:
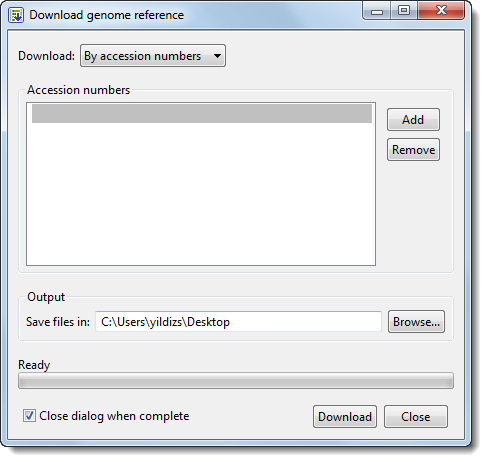
1) If you know the accession numbers for the templates you wish to download, choose By Accession numbers from the Download menu. The following version of the dialog is displayed.

2) In the Accession numbers section, type in the first template accession number. Press Enter or use the Add button to move to the next row, where you can add the next number. Continue adding accession numbers for all sequences you wish to use as templates. If you wish to remove a sequence from the list, highlight the sequence and click Remove.
3) Use the Browse button to specify a file location in which to save the templates.
4) If you need to add additional accession numbers as templates, uncheck the box Close dialog when complete.
5) Click Download to initiate download of the specified Accession numbers from NCBI. ArrayStar will automatically import the downloaded sequences as templates.
Downloading a whole genome:
1) If you wish to download an entire genome, choose Whole genome (the default) from the Download menu. The dialog updates to offer new options.
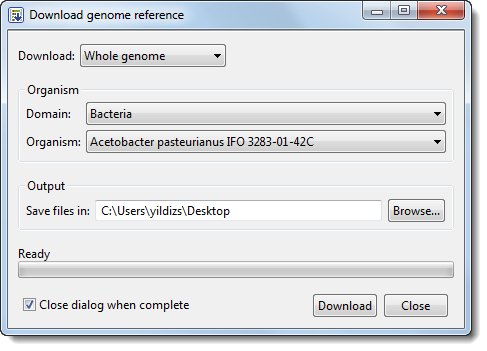
2) From the Domain menu, choose either Archaea, Bacteria, Eukaryota or Viruses.
3) From the Organism menu, choose the species name.
•If you do not see the genome you wish to download: Search the NCBI Genome database to see if the genome has been removed, or if it is listed under an alias. For example, searching for Ashbya gossypii will automatically redirect to its alias, Eremothecium gossypii, which you can then select from the Organism menu.
Some species have different genome builds available; in which case the species name is followed by a number. For species without a specific build number, ArrayStar will retrieve the most recent build of the selected genome.
•If you need to download an older version of a genome: One case in which you may wish to use an older version of a genome is when comparing new data with data that have already been analyzed using the older genome. Another possibility is that your reads are in a supported alignment file that uses an older genome’s coordinate system. To find and download an older version of a genome, search the NCBI Assembly database for the organism of interest. From the results list, click on the hyperlink for the genome version you wish to use. The ensuing page will contain a table listing individual chromosomes and their GenBank IDs.
4) Use the Browse button to specify a file location in which to save the templates.
5) If you need to add additional genomes as templates, uncheck the box Close dialog when complete.
6) Click Download to initiate download of the specified genome. ArrayStar will download all the reference sequences for the selected genome from the NCBI Entrez Genome Project database. These downloads may include auxiliary genomes such as mitochondria and chloroplasts. They may also include some contigs which have not yet been placed by the genome finishing process.
Note: ArrayStar uses NCBI’s eUtils service to download Entrez sequence files. Click for more information about eUtils or to read NCBI’s Disclaimer and Copyright notice.