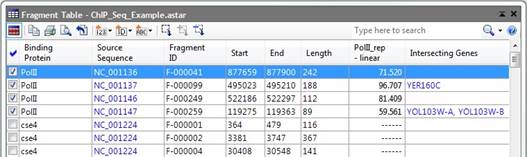
The Fragment Table shows detailed information about the number of reads that map to different fragments of the genome in ChIP-Seq and miRNA experiments.

During processing, QSeq will assign a “Fragment ID” to each section of the template sequence where reads are mapped. By default, all of the fragments in your project assigned “Fragment IDs” will be displayed each time you open a new Fragment Table. The Fragment Table may be used to show all of the fragments in your project, or only the currently selected fragments. Various fields, including a variety of annotation fields, notes, and signal values can be displayed in the Fragment Table. Some annotation values include genes that intersect or are upstream or downstream of the fragment. Fragment expression values correspond to how many reads are mapped to a given fragment of the template.
For more information, see the following help topics:
•The availability of tables in different workflows.
•Descriptions of columns that can be added to the tables.