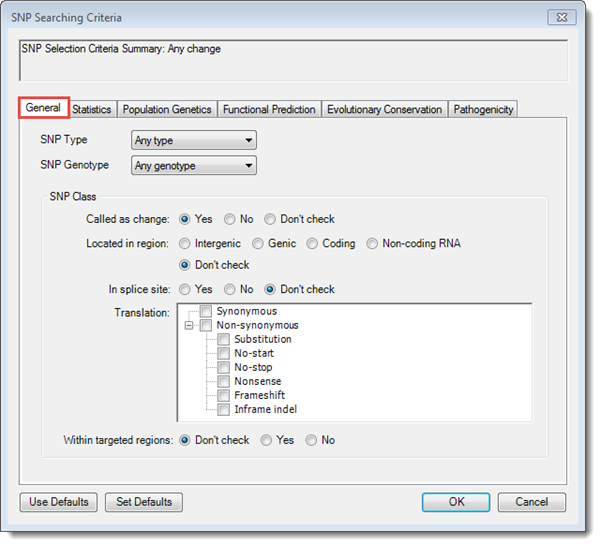
The tabs in the SNP Searching Criteria dialog control which variants will be displayed in the SNP Table. For projects imported from SeqMan NGen, the General tab is populated with the same choices that were set up prior to assembly.

•SNP Type –Use the dropdown menu to choose between SNPs only, Indels only, or Any type.
•SNP Genotype – Use the dropdown menu to choose between seven categories (e.g. Homozygous Variant, Any Heterozygous, etc.)
•In the SNP Class section, chose from options in the subsections Called as change, Located in region, and in splice site. The first two subsections are grayed-out if any boxes are checked in the Translation section below.
•Translation – Check the boxes to select one or more translation change types:
|
Type |
Description |
|
Synonymous |
This change does not alter the amino acid sequence coded for in this (translated) coding region. |
|
Non Synonymous |
This change alters the amino acid sequence coded for in this (translated) coding region. This category includes all types below it. To derive the number of simple substitutions, display all of the options and subtract the four listed changes from the Non-synonymous count. |
|
Substitution |
A single-nucleotide polymorphism (SNP). This category is a summation of SNPs from each individual category (e.g. Nonsense, No-stop, etc.). Only substitutions are included in the count; indels are excluded. |
|
No Start |
This change results in absence of a start codon. |
|
No Stop |
This change results in absence of a stop codon, thereby extending the reading frame. |
|
Nonsense |
This change results in a premature stop codon and a truncated, incomplete, and usually nonfunctional protein product. |
|
Frameshift |
An indel within a coding region and which is not a multiple of 3, thereby changing the reading frame. |
|
Inframe Indel |
An insertion or deletion within a coding region whose length is divisible by 3.
•For insertions, the type will be followed by the word Conservative when inserted bases occur between two codons, and by the word Disruptive when the inserted bases occur within a codon.
•For deletions, . the type will be followed by the word Conservative when deleted bases begin at the first position of a codon and end at the last position of a codon, and by the word Disruptive when the deleted bases start at position 2 or 3 of a codon and end at position 1 or 2, respectively, of another codon. |
•Within targeted regions – If you are using a targeted region (BED or manifest) file, choose Yes to include only those SNPs that are present in the targeted regions, or No to include only those which are absent. Choose Don’t check to include both types of SNPs.