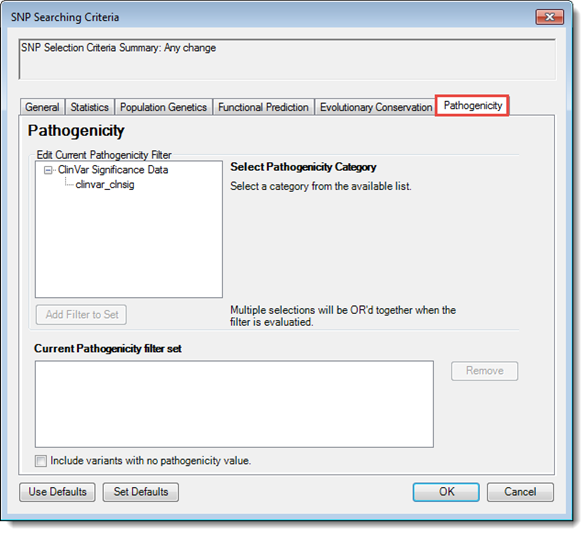
Each of the tabs in the SNP Searching Criteria dialog control which variants will be displayed in the SNP Table. The Pathogenicity tab lets you filter based on predicted pathogenicity. Unannotated variants are excluded from this filter.

1) Expand the Current Pathogenicity Filter tree, as needed, until you see the database of interest. For descriptions of each category, see Table Column Descriptions.
2) Make a selection from the tree. The name of the pathogenicity category and its description appear to the right
3) Use the drop-down menu to select a Boolean expression. To the right of the menu, check one or more boxes corresponding to the desired result categories. For example, the selections made in the image below would display only those alleles with a ClinVar pathogenicity score indicating that the variation is likely not due to histocompatibility.
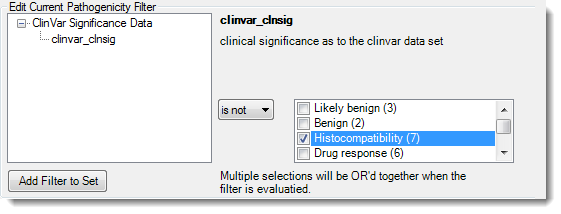
4) Once you have chosen the desired options, click Add Filter to Set. The filter set will appear in the box in the lower half of the dialog.
5) (optional) To add a filter for another population, or using different criteria, repeat steps 1-4. If you need to remove a set from the lower part of the dialog, select it from the list and click the Remove button.
Example: To find all deleterious variants, you could set up two filters: “is not” + “Benign” and “is not” + “Likely benign.”
6) To include novel SNPs in the search (i.e., SNPs for which no pathogenicity information is available), check the box Include variants with no pathogenicity value.