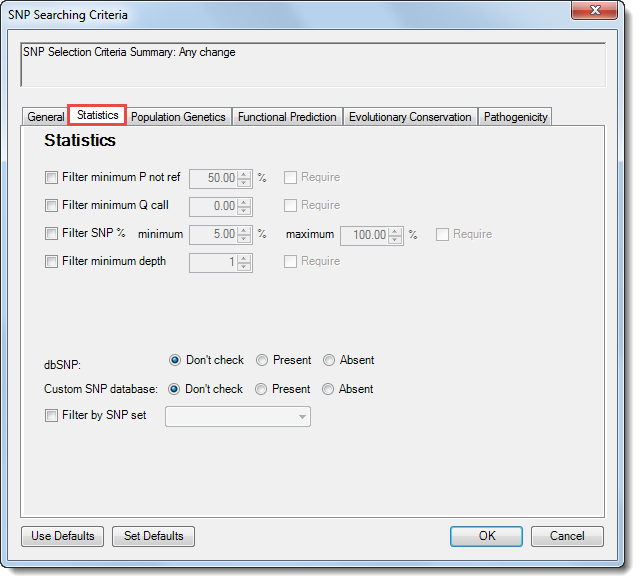
The tabs in the SNP Searching Criteria dialog control which variants will be displayed in the SNP Table. The Statistics tab allows you to filter based on statistics such as depth or the probability that the variant is not the same as the reference sequence; as well as presence of the variant in the Database of Single Nucleotide Polymorphism (dbSNP) or your custom SNP database.

•The first four rows allow you to filter based on statistics or depth of coverage.
|
Filter minimum P not ref |
The probability that this position does not match the reference. For combined SNPs and indels, P not ref will be the minimum of the P not refs in the used columns. |
|
Filter minimum Q call |
The minimum Q call, where Q call is a measure of the confidence that the SNP is present in the sample on a log10 scale of 0-60. Q call is similar to the Phred quality score of the called genotype. For combined SNPs and indels, Q call is the minimum of all available columns at that reference position. |
|
Filter SNP% |
The percentage of the sequence at this position in the assembly which varied from the reference. If this is selected, you may enter both minimum and maximum threshold values. |
|
Filter minimum depth |
The minimum depth of coverage for the SNP. |
Check the corresponding Require box only if you want ArrayStar to disregard all data that do not both contain the necessary information and meet the threshold requirement
Example: You import an .assembly file which contains SNP, P not ref, and minimum depth data. You also import a VCF file that contains SNP and minimum depth data but lacks P not ref calls. You then filter for any change in which P not ref is ≥ 0.90 and minimum depth is ≥ 10.
o If Require is unchecked, ArrayStar locates any positions that match all three criteria in the .assembly file and the two criteria that are present in the VCF file.
o If Require is checked, ArrayStar requires that all three criteria be fulfilled in both experiments, and the filter will thus result in zero matches.
•Filter GERP scores – If this option is enabled, you may check the box and use the two drop-down menus to filter data that is ≤ or ≥ the chosen GERP score. SNP positions with GERP scores less than 1 are not included in the genome template package. An empty cell in the GERP column indicates that the score is less than 1.
•dbSNP – If Database of Single Nucleotide Polymorphism (dbSNP) information is included in the assembly data, you may choose Present to include only those SNPs that are present in the table. Choose Don’t check to include both present and absent SNPs.
•Custom SNP database – If you are using a VCF SNP file, you may choose Present to include only those SNPs that are present in the table. Choose Don’t check to include both present and absent SNPs.
•Filter by SNP Set - Check this box and use the drop-down menu to choose from sets created in the Set List. This search option is only displayed if you selected “Genes” in the Search for section of the Advanced Filtering dialog.