If you are following the Variants workflow, the Set Up Preprocessing step of the Project Setup Wizard contains an Advanced Options button that allows you to review and edit advanced pre-processing options. Clicking this button leads to a multi-tabbed dialog.
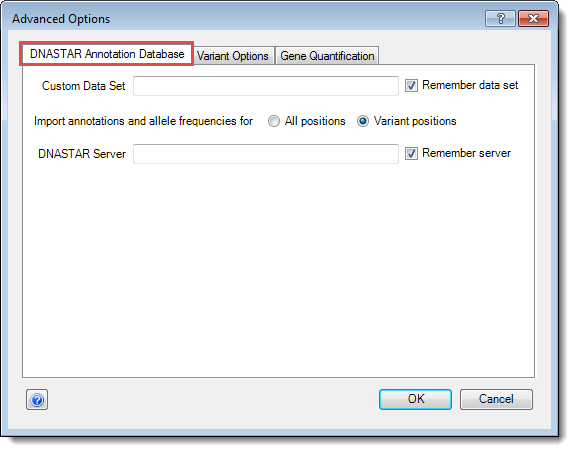
The DNASTAR Annotation Database tab:

•If you wish to use a Custom Data Set, enter its name in the text box provided. To ensure the same set is automatically entered for future ArrayStar projects, check the box next to Remember data set.
•Under Import annotations and allele frequencies, choose the positions for which database information should be retrieved:
o Choose All positions to include reference calls as well as variants.
o Choose Variant positions to include only positions called as variants.
•To use a specific DNASTAR Server, enter its web address in the text box. To confirm the same server should be automatically entered for future ArrayStar projects, check the box next to Remember server.
The Variant Options tab:
This tab is available to all users and contains the majority of available advanced options.

•Under “What kind of variant is required…,” choose whether you want to load SNPs with Any change, or those with Any non-synonymous change only.
•Under “How many experiments…,” enter the Minimum # experiments that must have a change at the same position before SNPs are loaded. In addition, you can use the optional checkboxes to specify that ArrayStar Ignore the minimum number if there is a non-synonymous and/or gene-disrupting change.
•Under “Import additional information…,” use the optional checkboxes to import additional information about selected experiments. Both options only import reference positions where useful SNPs are present.
o Locations noted in dbSNP – Imports Database of Single Nucleotide Polymorphism (dbSNP)table information from a SeqMan NGen project.
o Custom user SNP positions – Imports the custom VCF SNP table from a SeqMan NGen project.
•Under “Advanced criteria for choosing positions…,” specify which type of SNPs should be imported. Three choices are available.
o Always include all available positions – In Variants projects, called SNP information is loaded by default. Checking this option also loads information on reference call positions (from dbSNP or a VCF file) in the .assembly file, whether or not a variant has been called in one of the project samples. Note that if this box is checked, several of the subsequent options are disabled.
o Always include all custom user SNP positions – To load information from each position in the custom user VCF file, within the selected region, whether or not that position has been called a variant in any sample. Positions from other sources (e.g., dbSNP) that were called the reference in the assembly are not loaded into the ArrayStar project. To enable this option, uncheck Always include all available positions.
o Only include custom user SNP positions – To load only positions in the custom user VCF file that are both called as a variant in at least one sample and are within the selected region. To enable this option, uncheck Always include all available positions.
The Gene Quantification tab:
This tab is available to all users and contains only options related to gene quantification. Gene Quantification is used to generate a numeric “signal” for protein encoding genes only and is meant to reflect the cumulative effect of all called variants on the protein. It is the value ArrayStar uses for scatter plots and clustering algorithms.
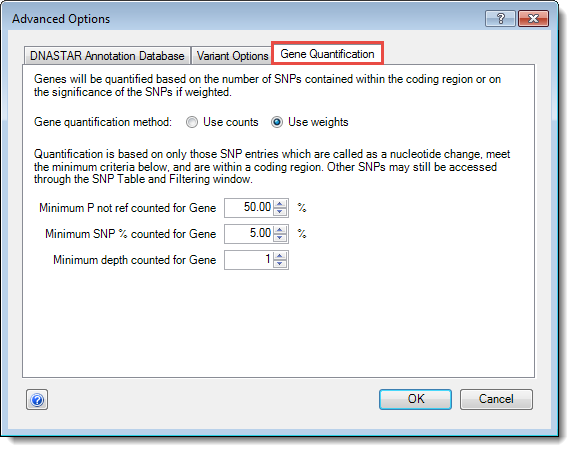
•To the right of Gene quantification method, choose whether to use counts or weight in preprocessing:
o Use counts – Determines a “signal” value for each gene by simply counting the number of SNPs and indels.
o Use weights – Determines a “signal” value for each gene using a weighted scoring system where each synonymous SNP is given a value of 1, each non-synonymous SNP resulting only in an amino acid substitution is given a value of 100, and each nonsense or frameshift mutation is given a value of 10,000. Heterozygous SNPs are given a halved weight (0.5, 50, or 5000). The weighted scores are then added together to give the final signal.
•You can filter the number of variants used for the gene quantification using any combination of the following three parameters.
Note: These filters only apply to calculation of the gene quantification signal and will not affect the presence or absence of a variant elsewhere in the project.
o Minimum P not ref counted for Gene – The minimum probability that the called base (or multiple bases) at a position is not the reference base. Default is ≥ 50%.
o Minimum SNP% counted for Gene – The minimum percentage of reads in which the SNP must occur. Default is ≥ 5%.
Note: Depending on your selections for Minimum SNP % and Minimum P not ref, there is a slight possibility of a "rounding error" occurring, in which the Gene Table may report one thing (e.g., an experiment has no Nonsense genes) whereas the SNP Table for that gene shows contradictory information (e.g., the experiment has a Nonsense gene).
o Minimum depth counted for Gene – The minimum depth of coverage at a position for that position to be considered.
After making the desired changes in all tabs present, click OK to save your changes and return to the Set Up Preprocessing screen, or Cancel to return without saving changes.