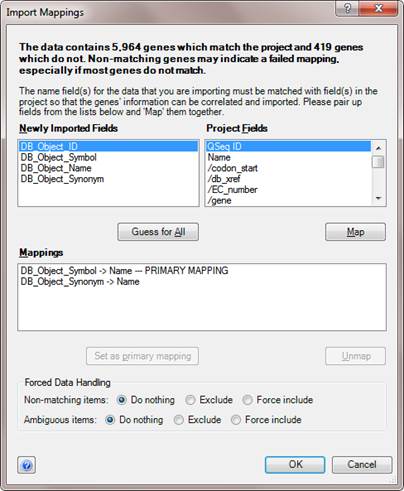
During file import mapping, clicking the Advanced button launches the Import Mappings dialog. This dialog also appears when you are importing new experiments with non-matching or ambiguous fields. Import Mappings allows you to manually provide a better mapping of fields.

The Newly Imported Fields section displays the fields from the file(s) being imported.
The Project Fields section displays the fields that are already within the project, or the fields from the batch of files being imported, if automatic mapping is not possible when a batch of files is being imported together.
To map fields automatically:
To attempt automated mapping, click the Guess for All button. If ArrayStar can guess, the two fields will be listed in the Mappings section of the dialog, to indicate that they have been paired.
If ArrayStar cannot guess, you will receive the following error message:
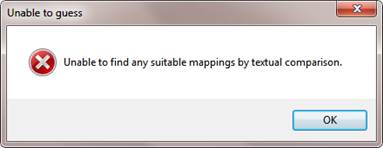
Click OK, then proceed using the directions under "To map fields manually."
To map fields manually:
To map fields manually, select one field from each list, choosing the fields that contain the gene names that should be used to uniquely identify each gene. After doing so, click the Map button. The two fields will be listed in the Mappings section of the dialog, to indicate that they have been paired.
In addition to pairing fields to create mappings, the Import Mappings dialog also allows you to specify one mapping as the primary mapping by selecting one and choosing Set as primary mapping. This causes other fields in the file to be ignored when mapping the values within these fields, unless there are genes within the two fields that are duplicate or ambiguous.
The Forced Data Handling options allow you to specify how ArrayStar will treat non-matching and ambiguous fields in the imported file. You may choose from the following options:
•Do Nothing – ArrayStar will attempt to automatically include new items when it is safe to do so without requesting further input from you. For example, ArrayStar considers it safe to import new items into an empty project. but unsafe to import many new items into a project that is already populated with experiments or annotations. If you choose this option, the Import Mappings dialog will appear again if ArrayStar considers it unsafe to import ambiguous or non-matching items.
•Exclude – All non-matching or ambiguous items will be excluded from the project so that only the unambiguously matching items are imported. This is often useful when importing annotations to avoid having genes created which are unrelated to your data set.
•Force Include – Non-matching or ambiguous items will be included in the project. For non-matching items, this will mean including fields that do not map to anything in your project, resulting in blank entries in various columns of the Gene Table. When importing annotations that match multiple genes, imported terms will be applied to all items in the project which they match. When importing experiments with ambiguous fields, the signals and accompanying annotations will be placed in new entries to avoid any duplicate signal values.
Once you are finished specifying mappings, click OK to proceed with importing.