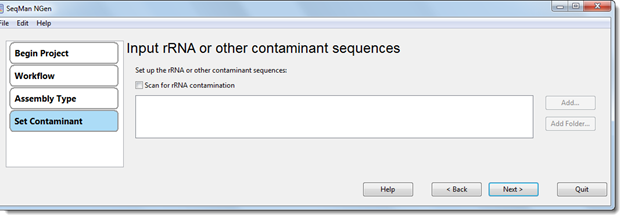
If you are following the transcript annotation workflow (de novo Transcriptome/RNA-Seq), you may enter the rRNA sequences here in order to screen out contaminants during assembly.
Note: It is not necessary to enter anything in this screen; if desired, you may press Next to continue to the next wizard screen.

•Scan for rRNA contamination – Check this box to enable the three add/remove sequence buttons on the right. You will then need to specify rRNA sequences here, as described below.
To add files or folders of files:
•Local assemblies - Add files (e.g., FASTA or Genbank files) using the Add button or add folders of files using the Add Folder button. In the file explorer, navigate to and select the desired file(s)/folder(s), and then click Open.
•Cloud assemblies - Add files or folders using the Add button. This
takes you to the Cloud Data Drive. Navigate to the desired file(s) or folder and
then click the green check mark ( ).
).
To remove files:
If you would like to remove a file from the list, select it and click Remove.
***************
Once you are finished making choices in this screen, click Next > to continue to the next wizard screen.