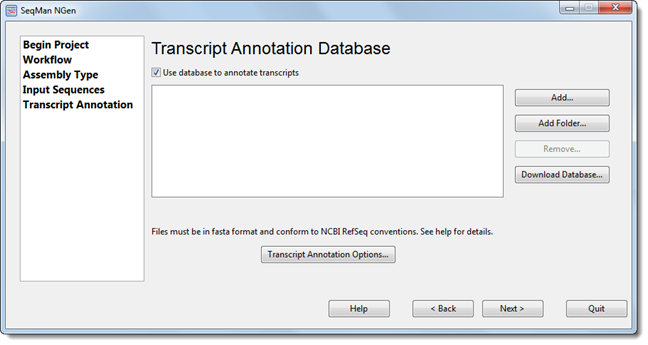
The Transcript Annotation Database screen appears only as part of the transcript annotation (i.e., de novo Transcriptome/RNA-Seq) workflow.

Leave Use database to annotate transcripts checked if you wish to annotate transcripts during assembly using information from a specified database. You will need to specify a transcript annotation database, as described below.
If you prefer to uncheck the box, all other buttons in the dialog will be disabled; in that case, press Next > to proceed to the next screen.
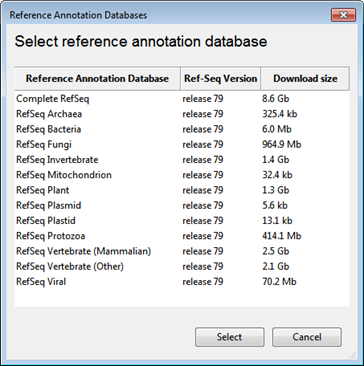
To download one or more databases from the DNASTAR website:
Licensed users can access DNASTAR’s database of transcript annotations extracted from data on NCBI’s RefSeq website.
From the Transcript Annotation Database of the SeqMan NGen wizard, click the Download Database button. Select one or more items from the list and then press Select.
To use a database from your local computer (local assemblies only):
Add files using the Add button or add folders of files using the Add Folder button. Use the file explorer to navigate to the desired file(s) and then click Open. The file must meet strict formatting standards, as described in Creating a Custom Transcription Annotation Database.
To use a database from the DNASTAR Cloud Data Drive (Cloud assemblies only):
Add files or folders using the Add button. This takes
you to the Cloud Data Drive. Navigate to the desired file(s) or folder and then
click the green check mark ( ).
).
Note: When using the Add or Add Folder buttons, the files being added must conform to strict formatting specifications and must be in .fasta format.
To remove a database from the list:
Select the database you want to remove and click Remove.
*****************
To access advanced options, press the Transcript Annotation Options button.
When you are finished with the Transcript Annotation Database screen, click Next > to proceed to the next wizard screen.