Note: This topic is not applicable to BAM-based projects.
SeqMan Pro can identify any reads containing repetitive sequences prior to assembly. SeqMan Pro will check each added sequence for matches to the specified Repetitive Sequences. If a match is found, the matching sequences are added last to the project to reduce the chance that repetitive sequences will “poison” the assembly with inappropriately assembled reads.
Note: Also see the Use Repeat Handling parameter under Assembling parameters.
To specify repetitive sequences:
1) Store sequence files of repetitive sequences in the Repetitive Seqs folder, found in the following location:
Windows 7 & Windows 8: C:\Users\Public\Public Documents\DNASTAR\Lasergene 9 Data\
Macintosh: Hard Drive:Applications:DNASTAR:Lasergene
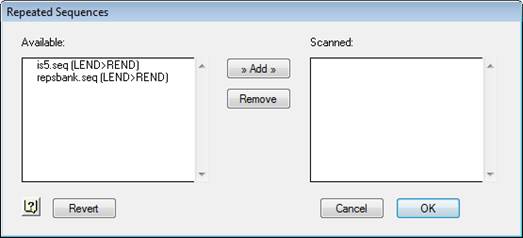
2) Select Project > Repetitive Sequences. Files stored in the Repetitive Seqs folder appear in Available on the left.

3) Specify repetitive sequences by selecting the appropriate sequences from the list on the left and clicking Add.
Selected sequences move to the Scanned list.
If you decide not to use one of these sequences, highlight its name in the right window and click Remove.
4) When you’ve specified all of the repetitive sequences, click OK.
To activate the handling of repetitive sequences:
1) Add sequences to the Unassembled Sequences window.
2) Click the Options button to open the Preassembly and Assembly Options dialog.
3) Check Remove contaminant sequences and Optimize Sequence Assembly Order.
4) Finally, choose one of the following options:
•Click Scan Selections to search for repetitive sequence in highlighted sequences only.
•Click Scan All to search all sequences.
•Click Scan Later to postpone searching until clicking Assemble. Upon scanning the sequence, repetitive sequences will be located and the reads that contain them will be ordered for addition to the assembly last.