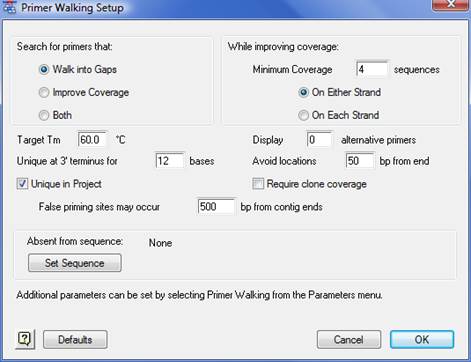
The Primer Walking Setup dialog allows you to select the parameters for SeqMan Pro to use when choosing primer locations. Access the dialog by selecting Contig > Primer Walk.

Adjust the parameters in the Setup dialog as needed:
•Walk into Gaps - Selects a search that primes locations at the ends of contigs, polymerizing into the gaps between them. The primers are selected individually and assumed to amplify linearly.
•Improve Coverage - Selects a search that primes locations that surround areas of low coverage. The primers are selected individually and assumed to amplify linearly.
•Both - Combines Walk into Gaps and Improve Coverage search options.
•Minimum Coverage - Determines the upper bound on locations that will be considered for coverage improvement. Areas that are deeper than the minimum will be excluded from the search.
•On Either Strand/On Each Strand - Applies the minimum coverage value to the overall depth of the contig (Either Strand) or applies it to the minimum of the depth on one strand (Each Strand).
•Target Tm - Represents the value for melting temperature that will become the target for the entire set of results. Optimal primers will be chosen as close to this value as possible, provided they meet the other constraints. Primers with melting temperatures far from the target value are weighed lower linearly by temperature difference. Widening the range is the first response toward heuristic resolution.
•Display ‘n’ alternative primers - Increasing this value causes the search algorithm to present additional primers whose role in extension or coverage overlaps the most optimal choice. The number of primers presented for a given purpose is equal to this value plus one.
•Unique at 3’ terminus for ‘n’ bases - Represents the length that defines uniqueness for 3’ dimerization with the template. The default value is raised to correlate with the total amount of sequence in the project.
•Avoid locations ‘n’ bp from end - Weights the location of primers maximally at this distance from the contig ends, tapering linearly in either direction. The value overrides the confidence in the quality of sequence reads near their ends forcing greater overlaps when the value is increased.
•Unique in project - When checked, it causes primers that occur in more than one location within the entire set of contig consensus to be excluded. Contigs composed of a single sequence are included. Primers that share more than one oligo at 3’ of the length are excluded.
•Require clone coverage - When checked, it forces primers to be considered only when at least one sequence read overlaps the primer location on the strand being primed.
•False priming sites may occur ‘n’ bp from ends - Represents the value that places a limit on the locations that we are concerned about when false priming sites are found. Sites more toward the middle of contigs than the specified value are presented, but not counted toward the rejection of a primer for end extension.
•Set Sequence button - Allows you to supply the sequence of a vector or contaminant which must not prime a polymerization. Primers that share an oligo at 3’ of the length are excluded.
Click OK. The results of the primer walk are displayed in the Strategy View, as well as in report format by selecting the Primer Walking Report.