Note: This topic is not applicable to BAM-based projects.
SeqMan Pro can remove contaminant sequences during the preassembly process. For example, if you are sequencing yeast DNA inserts that were cloned in E. coli, you may want to remove any contaminating E. coli DNA from the project prior to assembly.
Sequence files of known contaminants should be stored in the Contaminant Seqs folder found in the following location:
•Windows 7 & Windows 8: C:\Users\Public\Public Documents\DNASTAR\Lasergene 9 Data
•Macintosh: Hard Drive:Applications:DNASTAR:Lasergene
Note: If desired you may create a File of Filenames for a group of contaminants and then save it in the Contaminant Seqs folder. This can be useful if you want to specify only a portion of the contaminant sequences to scan.
To remove contaminant sequences from your assembly:
1) Store the contaminant sequence(s) in the appropriate directory list above.
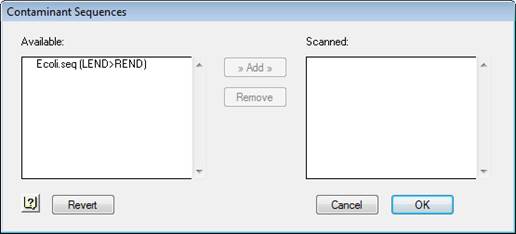
2) Select Project > Contaminant Sequences. Files stored in the Contaminant Seqs folder will appear in the Available window on the left.

3) Specify known contaminants by selecting the appropriate sequences from the list on the left and clicking Add.
Selected contaminant sequences move to the Scanned list.
If you decide not to use one of these contaminants, highlight its name in the right window and click Remove.
1) Click OK to save changes.
2) Add sequences to the Unassembled Sequences window.
3) Click the Options button to access the Preassembly and Assembly Options dialog.
4) Select Remove Contaminant Sequences, then choose from one of the following options:
Click Scan Selections to search for contaminants against highlighted sequences only.
Click Scan All to search all sequences.
Click Scan Later to postpone contaminant removal until clicking Assemble.
Upon scanning the sequence, contaminants will be removed using the specified Contaminant Screening parameters.
Note: Unlike vector trimming, which eliminates only the segments of reads that match vector sequence, contaminant screening prevents the whole read from entering the project. You should therefore use this feature cautiously. If your project is medium in size (large projects should be assembled with SeqMan NGen), you may want to increase the minimum number of matches to ensure that chance similarities between contaminant and target sequences do not eliminate informative data from your assembly. Alternatively, you may turn off contaminant screening altogether, and be alert for contigs that comprise contaminant sequence.