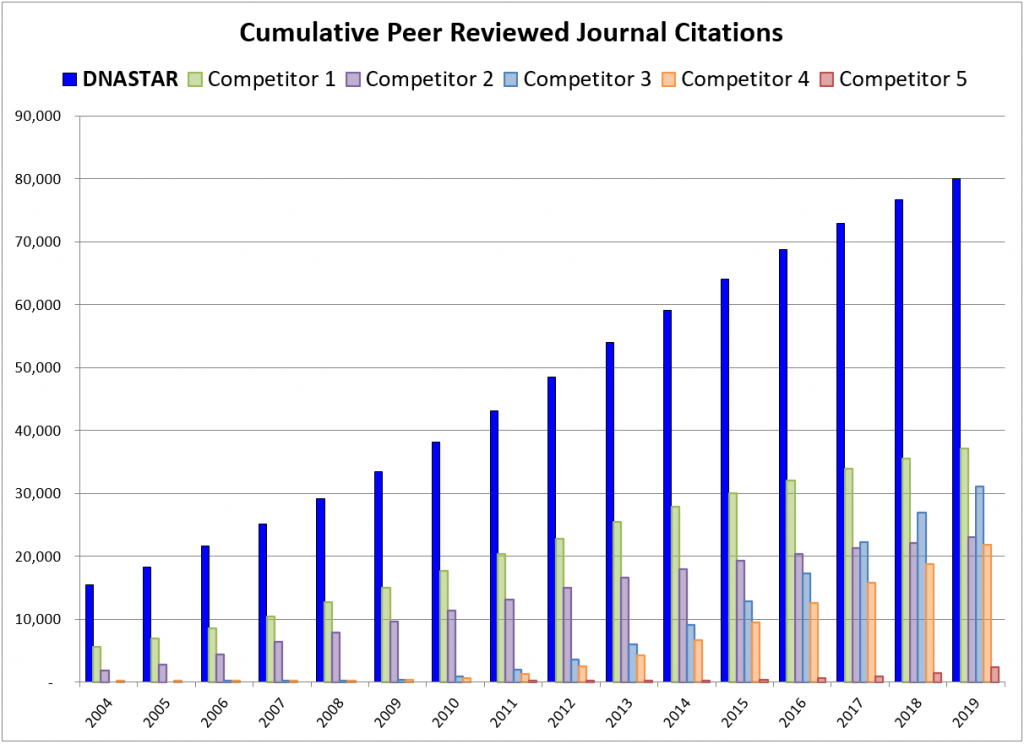
Over the past 35 years, DNASTAR software has been cited in over 80,000 peer-reviewed publications – more than any other sequence analysis software on the market. In 2019 alone, we were referenced in 3,370 peer-reviewed journal articles. We are grateful to consistently be the top choice in sequence analysis software to fulfill your research needs.
Examples of these publications include studies that used DNASTAR software for RNA-Seq analysis in Mexican cavefish; whole exome sequencing analysis for investigating developmental abnormalities in populations; and protein structure prediction to understand the role of HINT1 mutants in human motor disease, just to name a few. Other specific publications are highlighted in the Citations section of each of our new workflow pages.
Explore recent publications using DNASTAR software for different workflows:
- Virtual Cloning
- Sanger Sequence Assembly
- Multiple Sequence Alignment
- Pairwise Sequence Alignment
- PCR Primer Design
- Whole Genome and Whole Exome Sequencing Analysis
- RNA-Seq Alignment and Analysis
- De Novo Genome Assembly
- Protein Structure Prediction
Are you using DNASTAR software in your research? Our Publications Page features a suggested citation format that makes software citations easy to create. We would also love to feature your publication on our website and promote it on our social media accounts. For consideration, contact dnastar-info@dnastar.com or leave a comment on this blog post.


Leave a Reply
Your email is safe with us.