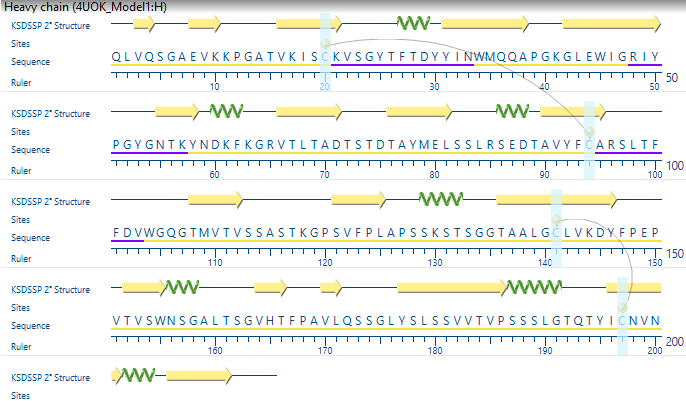
NovaFold Antibody
Creating accurate antibody models is crucial for antibody design and optimization. While many antibody modeling software tools rely solely on homology modeling, NovaFold Antibody’s unique algorithm generates models of antibodies and antibody fragments by combining homology modeling for framework regions and fragment-based or ab initio modeling for hypervariable loop regions, resulting in highly accurate antibody structure predictions. Simply provide sequences for the light chain, the heavy chain, or both, and NovaFold Antibody will return an annotated report that includes the predicted antibody models, as well as the highest likelihood heavy chain and light chain templates and annotations for locations of CDR loops. Easily view antibody modeling results with annotations in Protean 3D, part of Lasergene Protein, which allows you to fully customize the rendering to aid in visualization, or to prepare publication-quality images to export. Antibody modeling with NovaFold Antibody can also be used in conjunction with NovaDock to predict the structure and binding of antibody-antigen complexes.
Resources
Please see our resources below for more information on antibody structure prediction in NovaFold Antibody.
Antibody Modeling Workflow
How Can DNASTAR’s Protein Tools Help You?
Why Structure Prediction Matters
Nova Applications on Mac: Bringing the Power of Protein Modeling to the Desktop
High Resolution Protein and Antibody Modeling with NovaFold and NovaFold Antibody
NovaFold Antibody Help
FAQs
Do I need a Lasergene license to run NovaFold Antibody?
Yes. A Lasergene Protein license is required to generate antibody models in NovaFold Antibody. Protean 3D, included in Lasergene Protein, provides the interface for running NovaFold Antibody predictions and analyzing the results.
Do you offer a free trial of NovaFold Antibody?
One prediction for each of our Nova Applications is included in our Lasergene free trial. We also offer free, no-obligation, personalized demos of any of our Nova Applications with one of our scientists on staff, tailored to your specific research objectives.
What file types are supported for the light chain and heavy chain sequences required for antibody modeling?
Light chain and heavy chain sequences can be added in the following protein file formats: .aa, .fap, .fas, .fasta, .gp, .gbk, .sbd, .pro.
What is the prediction method used for antibody modeling by NovaFold Antibody?
The NovaFold Antibody algorithm utilizes a combination of homology modeling and ab initio loop prediction, resulting in highly accurate predictions. During modeling, NovaFold Antibody searches the input…
The NovaFold Antibody algorithm utilizes a combination of homology modeling and ab initio loop prediction, resulting in highly accurate predictions. During modeling, NovaFold Antibody searches the input sequence(s) against thousands of non-redundant protein antibody structures from PDB and finds the best template matches for the chain or complex.
During modeling, NovaFold Antibody gives particular consideration to the complementary determining region (CDR) loops, hypervariable regions of an antibody that react dominantly with an antigen. The three CDR loops on the heavy chain are known as H1, H2 and H3, while the three on the light chain are L1, L2 and L3. H3 is commonly the most important region in antigen binding. Due to its increased length and flexibility, it is also the most difficult to model. As such, NovaFold Antibody limits H3 loop modeling to fifteen or fewer residues, a length commonly seen in antibody modeling problems. Finally, NovaFold Antibody performs energy minimization calculations to construct the final predicted structure model for the antibody chain or complex. This entire process takes approximately 5-15 minutes on a standard workstation computer.