ArrayStar
Whether you are working with transcriptomics data to identify differential gene expression or performing large-scale variant analysis, sifting through vast amounts of NGS data can be cumbersome. ArrayStar, part of Lasergene Genomics, provides advanced filtering tools, robust statistical analyses, and rich, graphical views that allow you to effortlessly isolate gene or variant sets of interest and identify their biological significance. Explore gene ontology to discover how SNPs and small indels may impact gene function, or to identify relationships between genes with particular biological functions. If you work with human data, ArrayStar provides invaluable access to DNASTAR’s human variant annotation database, which integrates information from many large variant databases into your project for enriched variant analysis.

ArrayStar Workflows
Resources
Please see our resources below for more information on the tools available in ArrayStar.
7 Steps for Human Variant Analysis
Identifying Candidate Variants and Their Effects on Protein Structure Starting from NGS Data or VCF Files
Working with Variant Call Format Files in Lasergene Genomics
RNA-Seq Analysis using Lasergene Genomics
RNA-Seq Assembly and Normalization Methods—an Interview with Dr. Carl-Erik Tornqvist
Variant Analysis Webinar
De novo Transcriptome Assembly: SeqMan NGen vs. CLC Genomics Workbench
ArrayStar Help
ArrayStar Tutorials
The best!
“Gene expression analysis in Arraystar is the best!”
FAQs
Which Lasergene package includes ArrayStar?
ArrayStar is included in Lasergene Genomics as well as in the complete DNASTAR Lasergene package which includes all of the applications from Lasergene Molecular Biology, Lasergene Protein, and Lasergene Genomics.
What workflows are supported by ArrayStar?
ArrayStar provides in-depth gene expression and variant analysis, following an assembly of genomic or transcriptomic data in SeqMan NGen. As an analysis tool, ArrayStar supports clinical research studies, metagenomic assemblies, whole genome/exome sequencing, ChIP-Seq and RNA-Seq alignments, and de novo transcriptome analysis.
What types of variant analysis are available in ArrayStar?
ArrayStar provides a wide variety of tools for analyzing variants, including:
- Large-scale SNP comparisons across individuals and groups…
ArrayStar provides a wide variety of tools for analyzing variants, including:
- Large-scale SNP comparisons across individuals and groups
- Advanced gene filtering based on the level of disruption to each gene caused by variations
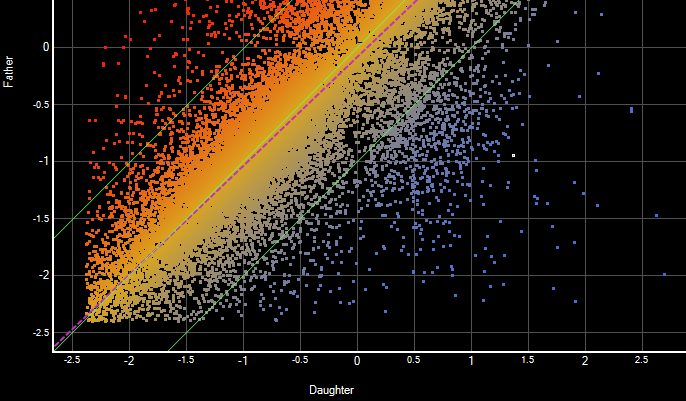
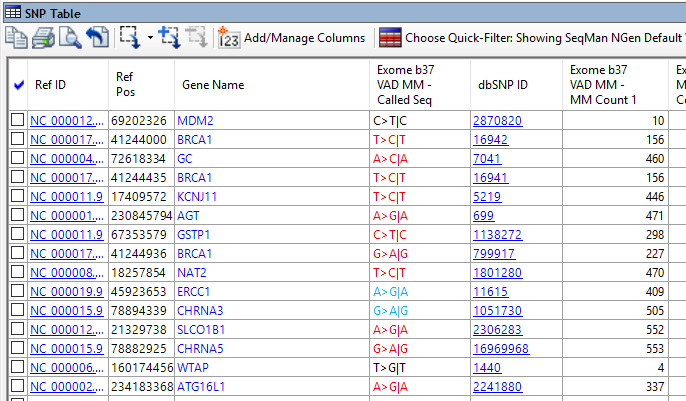
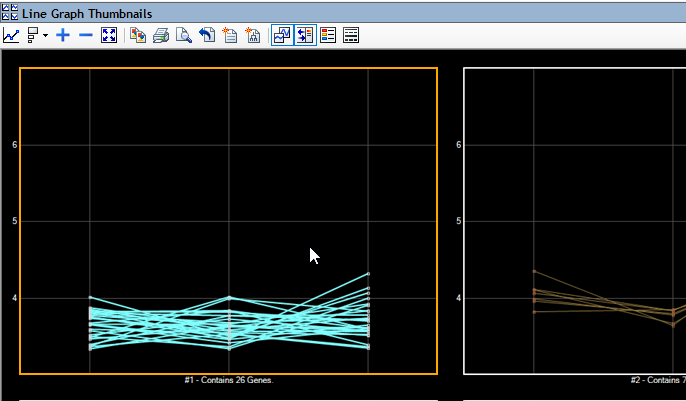
- Comparison of groups of variants using text filters, tabular data, or graphical representations that include Venn diagrams, scatter plots and heat maps
- The ability to send genes or variants of interest to SeqMan Ultra to view the read alignment at that position; or to GenVision Pro to view the assembly coverage for all samples simultaneously
What supplemental variant data is available in the Variant Annotation Database?
For human resequencing data, ArrayStar provides access to the Variant Annotation Database, which contains variant information using coordinates from GRCh37 (hg19) and GRCh38. Annotations include information about the frequency of the variant in the general population, in specific populations, and in publications, as well as information concerning the variant’s impact on functionality. The annotation information comes from several sources, including:
- Mastermind, the comprehensive database of genomic literature from Genomenon, providing access to NGS variant data from millions of PubMed publications.
- 1000 Genomes Project and the Exome Sequencing Project (ESP)
- dbNSFP v 4.1 which includes:
- Functional impact predictions from Sorting Intolerant from Tolerant (SIFT), MutationTaster, and the Likelihood Ratio Test (LRT).
- Evolutionary conservation scores from SiPhy, PhyloP, PhastCons and GERP++.
- Pathogenicity and clinical significance impact from ClinVar.
- Basic amino acid sequence information, Database of Single Nucleotide Polymorphism (dbSNP) IDs, and annotations from Uniprot and Interpro.
What types of RNA-Seq analysis options are available in ArrayStar?
Analyzing RNA-Seq data in ArrayStar enables you to combine gene expression data, isoform info, detected variants, annotations and ontology in a single project. You can compare gene expression values between experiments using scatter plots, heat maps, and line graphs. In addition, you can identify genes, isoforms and variants of interest through simple or advanced filtering based on signal, annotation values or many other options.
Compare DNASTAR Lasergene Packages
| MOST POPULARDNASTAR Lasergene | ||||
| Lasergene Molecular Biology | Lasergene Genomics | Lasergene Protein | ||
|---|---|---|---|---|
| Included Applications | ||||
| SeqBuilder Pro | ||||
| SeqMan Ultra | ||||
| MegAlign Pro | ||||
| GeneQuest | ||||
| GenVision | ||||
| SeqNinja | ||||
| SeqMan NGen | ||||
| ArrayStar | ||||
| GenVision Pro | ||||
| Protean 3D (+1 prediction per Nova Application) |
||||
| DNASTAR Navigator | ||||
| Supported Workflows | ||||
| Integrates with | ||||
| See Pricing | See Pricing | See Pricing | See Pricing | |