NovaFold AI & NovaFold AI-Multimer
Computational biology has come a long way in the past few years, and DNASTAR is leading the way in commercial protein structure prediction. Both the prediction and analysis take place within the existing NovaCloud Services interface of Protean 3D.
NovaFold AI handles even the most difficult single-chain folding problems
Our NovaFold AI protein structure prediction software enables you to seamlessly utilize the award-winning AlphaFold 2 method to predict the protein structure of an amino acid sequence, and then visualize and analyze your results. AlphaFold 2 was the top-ranked protein structure prediction method in the CASP14 challenge in 2020, significantly outperforming the other participating teams by determining the structure of proteins with accuracy comparable to laboratory experiments. In the most recent CASP Challenge, AlphaFold 2 achieved an average Z-score of 2.65 over 92 targets. The closest competitor achieved only 1.00 for the same number of targets. (See graph). AlphaFold 2 is extremely fast and typically finishes structure predictions in a few hours or less.
NovaFold AI is the first commercial software that can accurately predict even the most challenging protein structures:
• Membrane-bound proteins
• Fusion proteins
• Cytosolic domains (CDs)
• Extra-cellular regions
• G-protein-couple receptors (GPCRs)
NovaFold AI can also model multiple domains and their interactions with linkers, also known as multidomain protein structure prediction.

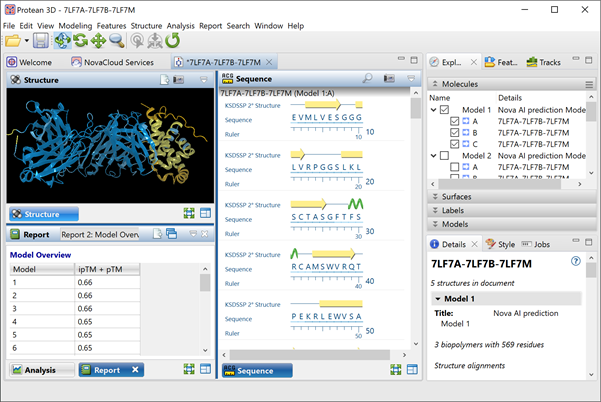
AlphaFold-Multimer has not entered CASP under its own name. However, the top three performers in the multimer target class of CASP15 (2022), the most recent experiment for which results have been calculated, used the algorithm as a foundational element of their approaches. For this reason, AlphaFold-Multimer is widely considered to be the most accurate algorithm to date for predicting the quaternary structures of protein-protein complexes.

Learn more about NovaFold AI and NovaFold AI-Multimer
Resources
Please see our resources below for more information on NovaFold AI powered by AlphaFold 2.
Protein Structure Prediction with NovaFold AI
Protein Structure Prediction with NovaFold AI-Multimer
Accurate Protein Structure Prediction
NovaFold AI Help
FAQs
Do I need a Lasergene license to run NovaFold AI protein structure prediction software?
Yes. A Lasergene Protein license is required to run NovaFold AI or NovaFold AI-Multimer. Protean 3D, included in Lasergene Protein, provides the interface for running predictions and analyzing the results.
Do you offer a free trial of NovaFold AI?
One prediction for each of our Nova Applications is included in our Lasergene free trial. We also offer free, no-obligation, personalized demos of any of our Nova Applications with one of our scientists on staff, tailored to your specific research objectives.
What protein folding prediction method does NovaFold AI use?
NovaFold AI uses the AlphaFold 2 artificial intelligence algorithm developed by DeepMind. Of over 140 prediction algorithms tested in the most recent CASP challenge, AlphaFold 2 was not only the most accurate, but was an astonishing 2.65 times more accurate than the next-best algorithm. AlphaFold 2 creates distance maps between residues using a graph neural network based on its training data of ~100,000 protein structures. Part of its prediction process is to compare to existing templates in the included library. Unlike NovaFold, which uses the I-TASSER algorithm, NovaFold AI does not directly use templates to place atomic coordinates. However, NovaFold AI does use templates as part of the map building process.
What protein folding prediction method does NovaFold AI-Multimer use?
NovaFold AI-Multimer uses the AlphaFold-Multimer artificial intelligence algorithm developed by DeepMind as an extension of its AlphaFold2 algorithm. It is optimized to predict multiple chain protein complexes, also known as “multimers.” AlphaFold-Multimer was introduced in 2021, one year after AlphaFold2.
To request access to NovaFold AI and/or NovaFold AI-Multimer, please complete the following form.


