Did you know that Lasergene–whether the licensed or the free trial version–comes with five free Cloud assemblies? Cloud assemblies are accessed through SeqMan NGen and allow you to run and manage next-gen assemblies directly on Amazon’s AWS cloud without tying up your valuable computer resources.
With DNASTAR Cloud Assemblies, you can run multiple NGS assemblies simultaneously, view assembly progress from any device, and readily access DNASTAR’s genome template packages for utilizing dbSNP, COSMIC, and GERP associations. DNASTAR Cloud Assemblies are compatible with all of our supported workflows, including whole genome sequencing, RNA-Seq, targeted resequencing, and more.
NGS Cloud Assembly and Analysis: Step by Step Instructions
If you don’t already have a license to Lasergene Genomics, download a free trial of our complete Lasergene package. You will have access to your 5 free NGS cloud assemblies as soon as you activate your software.
1) Open SeqMan NGen and choose New Assembly.
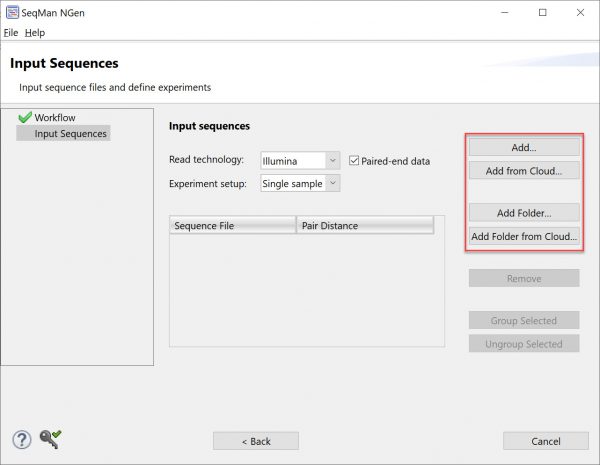
2) Follow the instructions within SeqMan NGen wizard to upload data and set up your project. You can upload your own data (using the Add or Add Folder buttons) or use DNASTAR demo data (using Add from Cloud or Add Folder from Cloud).
3) When you reach the Run Assembly Project screen, choose the option Run assembly on the cloud. Enter your DNASTAR login credentials if prompted.
4) Monitor the progress of the run by launching SeqMan NGen and choosing Manage Cloud Assemblies. When the assembly is complete, click on the Done link to download the results to your computer.
5) Analyze the sequence alignment, SNPs, and gene expression values on your local computer using SeqMan Ultra and/or ArrayStar.
If you need further help, we have step-by-step, illustrated tutorials, a 2-minute video SeqMan NGen Assembly on the Cloud, a 30-minute webinar Cloud Assemblies for NGS Sequences, and a help topic Cloud vs. Local Assembly to provide assistance.
Note that the five free assemblies allow you to assemble from 4-5 sequencing samples. For templated assemblies, one assembly is used for the QSeq normalization step, leaving you with four samples that can be assembled and analyzed in a single project. For de novo assemblies, each sample is equivalent to one assembly.
Need to assemble more samples?
Additional DNASTAR Cloud Assemblies are available to purchase online, with pricing available for both both academic/government institutes and commercial organizations.
Cloud Assembly Q&A with Matt Keyser
To get answers to some questions you might have about this service, we interviewed DNASTAR Senior Product Manager, Matthew Keyser.
What was the motivation behind developing the new Cloud Assemblies workflow?
DNASTAR assembly software is designed to work well on high-end desktop computers. However, laboratories are now moving toward lower-powered desktops or laptops. These computers often lack the memory (RAM) and/or hard disk performance (space and speed) needed for optimal assembly performance. Cloud Assemblies harnesses the power of the AWS cloud so that even very large assemblies can now easily be run from a basic laptop.
Are there advantages to using Cloud Assemblies vs. doing a local assembly on a powerful desktop computer?
There are several advantages to utilizing Cloud Assemblies rather than local hardware for NGS sequence assembly.
A cloud computer’s hardware can be more easily updated and optimized to accommodate specific alignment algorithms improving speed and performance and keeping costs down.
For example, the new NVMe SSD memory-based storage drives offer incredibly fast data throughput (>2500MBps), so that algorithms like SeqMan NGen, that frequently write to disk, can perform at a much faster speed. While it is possible to build local desktop computers with the same cutting-edge hardware, this approach often requires significant IT support to get all the hardware components working well together.
Cloud Assemblies also solves the challenge of storing and archiving both the raw input sequence data and the completed assemblies. With low overall cloud storage costs (initial costs, storage, maintenance, expanding storage), most users find cloud storage is far more cost-effective than an on-premise storage solution. Even better, DNASTAR provides free cloud storage to all Cloud Assemblies users!
Which workflows are better suited to local assembly vs. Cloud Assemblies, and vice versa?
If you consider the time needed to upload raw sequence data, assemble it, and download results, some projects are faster to run locally, while others are still much faster to run on the cloud.
For example, if you have at least a “medium-powered” computer, projects involving a small number of samples are often faster to run locally. As the number of samples increases, however, Cloud Assemblies jobs complete more quickly—even factoring in upload and download times—due to being able to run assembly jobs on multiple cloud computers simultaneously.
A single human RNA-seq assembly that takes two hours to complete on a powerful desktop may still take over an hour on a fast cloud computer. However, most RNA-seq experiments have multiple samples and replicates, and even a modestly-sized RNA-seq experiment of 10 samples will take 30+ hours to complete on a local computer. That same data set can be completed within a single workday (8-9 hours) using Cloud Assemblies.
In this situation, Cloud Assemblies can perform multiple assemblies in the time it would take to do a single assembly on a local computer.
Cloud Assemblies is also well suited to one-time large NGS sequence assembly projects that would not justify purchasing an expensive high-powered computer. For example, less common de novo transcriptome assemblies may require 32GB RAM and lots of hard disk space, and can often take 24+ hours to complete. Such assemblies are easy to do on the cloud and don’t tie up a local workstation either.
What about privacy and data security on the cloud?
DNASTAR Cloud Assemblies are powered by Amazon AWS, a secure, global cloud platform. For more information on cloud data security, please see our white paper DNASTAR Cloud Security.
With the growing amount of sequencing data being generated today, how do you see cloud solutions being used in the future?
I think the trend to using cloud computing for NGS sequence assembly will increase. As more powerful and less expensive cloud computing options become available, and as customer confidence in cloud security increases, it will become more difficult to rationalize purchasing and maintaining expensive local hardware solutions for assembly and storage. Utilizing cloud software provides users the best of both worlds: a friendly and familiar user interface for project setup and analysis, and access to powerful, cutting edge algorithms on the cloud.


Leave a Reply
Your email is safe with us.