Gene homology analysis allows us to better understand evolutionary relationships between organisms and to infer the functions of genes. Understanding sequence homology empowers scientists in a variety of fields, from archaeology and ecology to virology and drug discovery, to advance their research and make important connections from sequencing data.
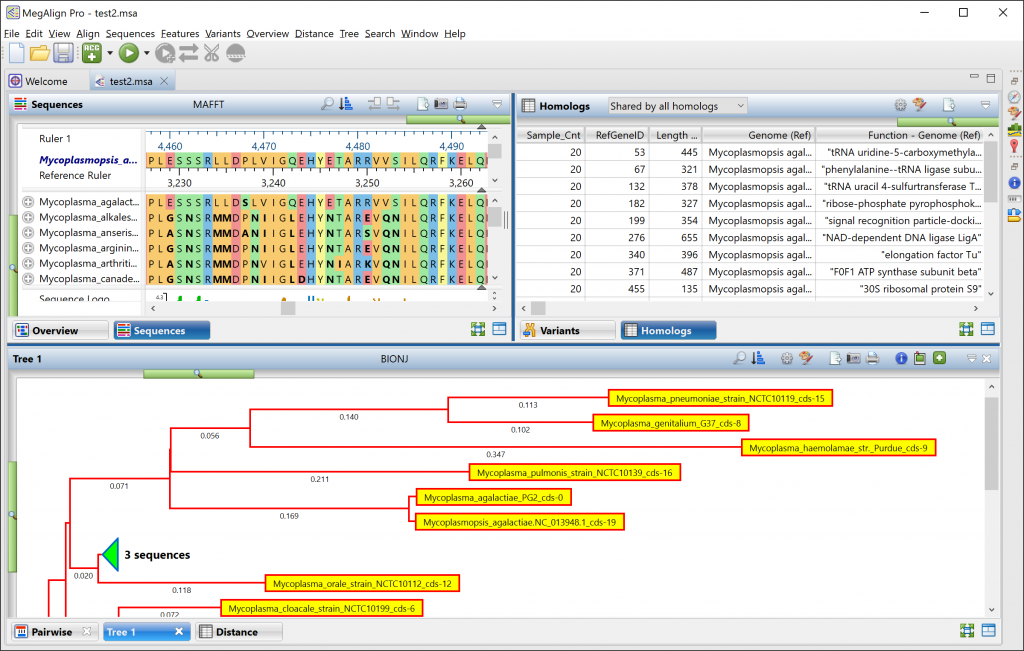
MegAlign Pro not only supports gene homology alignments for large data sets, including from whole chromosomes or entire bacterial genomes, but also presents the results in a way that clearly elucidates the evolutionary relationships between samples. The guided gene homology workflow in MegAlign Pro prompts you to select a reference sequence and upload sequence data for a bacterial genome or eukaryotic chromosome. Then MegAlign Pro identifies a subset of homologous genes and creates a concatenated protein sequence from each genome. The software automatically aligns these amino acid sequences against a reference sequence using the MAFFT alignment algorithm. The results include a phylogenetic tree and a comprehensive homologs table. Both of these can be fully customized with the data, layout and colors you prefer, and then exported as publication-quality images.
Sequence homology analysis in 4 simple steps
Step 1
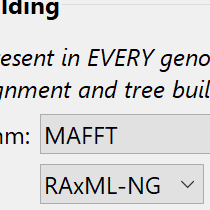
Specify sequences and alignment options

Step 2
Run the assembly
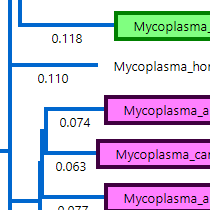
Step 3
Generate and compare phylogenetic trees

Step 4
View customizable tables of homologs
Resources
Please see the resources below for more information on performing sequence alignment and gene homology analysis with MegAlign Pro.
Webinar: Gene Homology at Scale
What Can We Learn from Gene Homology Analysis?
User Guide: Perform a Gene Homology Alignment
How this workflow was used to identify an unculturable bacterial strain in a hospital patient
Tutorials
Watch one of our videos or check out one of our written tutorials to learn more about using our sequence alignment software, MegAlign Pro.
Analyze Gene Homology with MegAlign Pro
Gene homology analysis can determine whether organisms share a common ancestor and is used in fields such as evolutionary and molecular biology, virology, ecology, pharmacy, and anthropology. Learn how to use MegAlign Pro to set up and run a gene homology alignment and analyze the results.
Adding Sequence Files for Alignment in MegAlign Pro
Learn how to load different nucleotide and protein sequences into MegAlign Pro for multiple and pairwise sequence alignment and phylogenetic trees. This video walks you through different ways to add and organize your sequence data prior to performing an alignment.
Supported File Types in MegAlign Pro
Learn about the different files types supported by MegAlign Pro, including FASTA files and GenBank files.
FAQs
What is the difference between gene homology or sequence homology and sequence similarity?
The link between sequence homology and sequence similarity (or identity) is often misunderstood. Simply put, sequence similarity indicates the percentage of similar residues between two sequences. By contrast, sequence homology is an inference drawn from the results of sequence similarity. Genes that share a common ancestor are referred to as homologs. To learn more, check out this blog post.
What methods does MegAlign Pro offer for computing phylogeny?
MegAlign Pro offers three methods for generating phylogenetic trees. The Neighbor Joining method uses the BIONJ algorithm. RAxML and its newer sibling, RAxML-NG are both used to create trees based on Maximum Likelihood calculations. RAxML-NG’s next generation algorithm is faster and has a larger capacity than its predecessor and is the default method for the gene homology analysis workflow.
Does MegAlign Pro support multiple phylogenetic trees in a single project?
Yes. MegAlign Pro makes it easy to have multiple trees for a single alignment, so that you can easily compare using different phylogenetic methods or changes to the alignment.
How do I export an image of my multiple sequence alignment?
Choose File > Export Image > (View Name) and choose the export format: Adobe PDF (.pdf), Microsoft PowerPoint-optimized (.pptx), PNG image (.png) or JPG image (.jpg, .jpeg).
Can I modify my multiple sequence alignment?
Lasergene’s sequence alignment software, MegAlign Pro, supports multiple ways to modify an alignment. You can:…
Lasergene’s sequence alignment software, MegAlign Pro, supports multiple ways to modify an alignment. You can:
- Trim ends at the beginning or end of an alignment.
- Realign a range of aligned sequences using different parameters or a different alignment engine.
- Merge two existing completed alignments together.
- Add all/selected sequences from the “Unaligned Sequences” area into the current alignment, while retaining all of the gaps of the original, pre-merge alignment.
- Realign existing aligned sequences plus selected unaligned sequences
Can I modify individual sequences within my multiple sequence alignment?
Yes. MegAlign Pro gives you the ability to reverse complement, edit, and trim individual sequences within an alignment.
What’s the easiest way to find sequence matches for my consensus sequence?
MegAlign Pro’s Search wizard, accessed using Search > Sequence Search, lets you search a query sequence against one of NCBI’s BLAST databases. The sequence can be part or all of an active sequence, another sequence file on your computer, or typed/pasted-in text.


