
Dr. Azzeh uses MegAlign Pro to study viral genetics in the Palestinian population
We recently interviewed virologist and DNASTAR customer, Maysa Azzeh about her work in and out of the lab. Dr. Azzeh has studied infectious viruses including hepatitis B, influenza A, and adenoviruses. Even before the current coronavirus outbreak, Dr. Azzeh’s work showed the true global nature of viral outbreaks and the importance of studying viral genetics. Her work on a viral database helped to verify potential outbreaks and may aid in vaccine development for Palestine and the surrounding region.
Dr. Azzeh’s friends describe her as resilient, determined, daring, and bold. These traits are reflected in both her research and outside hobbies. Her work on studying viral outbreaks is more relevant than ever, and she’s now finding new ways to help in her community during the current pandemic.
Could you give us a brief overview of your background and your career so far?
I work in the field of microbiology and virology with an emphasis on molecular biology and cell biology.
I was born in Palestine and grew up there. Since my childhood, I wanted to understand how we know things and to learn about the “invisible” world around us.
At the age of 18, I moved to Germany, where I lived until I was 30. As soon as I started reading about Microbiology, I knew this was the subject I want to study. After two years of general Biology and basic Sciences, I majored in Microbiology at Goethe University (JWG) in Frankfurt. I went on to earn my PhD and do my first post-doc in Germany before moving back to Jerusalem, where I did a second post-doc at the Hebrew University. I then established and served as the Head of the Virology Research Laboratory at Al-Quds University.
I moved from Jerusalem to NYC to work at the Touro College of Osteopathic Medicine as an Associate Professor in the Department of Basic Sciences. Touro is located in the heart of Harlem, just across the street from Apollo Theater on 125th Street. I am currently doing teaching and establishing a new research lab. During the current coronavirus health crisis, I am part of a health leadership group working in the local Harlem community.
What are some of your interests outside of your research?
Indoors, I enjoy aerobics and dancing. Outdoors, I love hiking, biking, and unicycling. My unicycle hockey team even competed at Unicon, the world unicycle convention. We were soundly defeated by the U.S. team, which turned out to include actual hockey players! I’ve been to over a dozen national and state parks in the USA, cycled through Moab, Utah, and hope to someday hike the 2,653-mile long Pacific Crest Trail. I also enjoy summer festivals; especially those with outdoor concerts.

How has your research evolved over time?
In 2007, I established the first and only Virology Research Laboratory (VRL) within the medical research center at Al-Quds University (AQU) in Palestine. I was assigned to direct the medical research center shortly after being recruited to AQU.
Until that time, I had been doing basic research studying the influence of cellular kinases on human cytomegalovirus (HCMV) assembly. Four students in my lab had focused their Masters’ theses on this research and I presented our joint research at the international Herpes Workshop (IHW).
Around 2009 I realized that the international science community had never researched the genetic epidemiology of circulating viruses in my country of Palestine. That’s when I started focusing on applied research in viral genetics.
Applied research at my lab started with a research project funded by the DFG-German Research Council. That study focused on the outcome of a new vaccine on children born to HBV positive mothers. I soon realized that nobody had investigated the circulating HBV genotypes among the Palestinian population. It had been assumed they were the same as those in surrounding countries. So, I began collecting HBV samples with the help of my students and also established a network with leading Palestinian physicians and hospitals.
At the same time, I had isolated Adenovirus from Palestinian children as part of an earlier project and had sequencing results waiting for analysis. My German partner in the DFG project advised me to buy DNASTAR software for my Virology lab. I purchased Lasergene in 2011 and my students and I started using MegAlign for sample analysis.
What are some of your research findings?
Scientific reports on infectious pathogens in the Palestinian population are rare. Genetic epidemiology data presented by my research contribute to the worldwide efforts to understand the epidemiology, distribution, and evolution of viruses. One of the goals of my research was to establish a database for a wide range of different viral genotypes. This database would be extremely useful as a public health tool for managing possible outbreaks and for designing vaccines for Palestine and the surrounding region.
For instance, after creating phylogenetic trees using DNASTAR software, we discovered that some viral sequences isolated from Palestinians residing in Palestine had a high similarity with viral isolates from the USA, Mexico, or Thailand. Even before the emergence of COVID-19, this emphasized to us that viruses know no borders.
In case of the influenza A publication (Bakri et. al., 2019), a major finding was that while genetic properties of Palestinian A(H3N2) and A(H1N1)pdm09 were in line with global isolates, A(H3N2), the dominant subtype in the northern hemisphere, was not dominant in Palestine. In addition, A(H1N1)pdm09 prevalence did not match World Health Organization findings for neighboring countries in the Middle East. We used both NCBI- and GISAID-archived influenza A sequences to place the Palestinian sequences into perspective. Some of our sequences showed higher percent identity with isolates from the USA than with other regional isolates.
Our research on HCV genotypes in Palestine (Rayan Da’as and Azzeh, 2019) showed a mix of regional influences rather than a single dominant genotype.
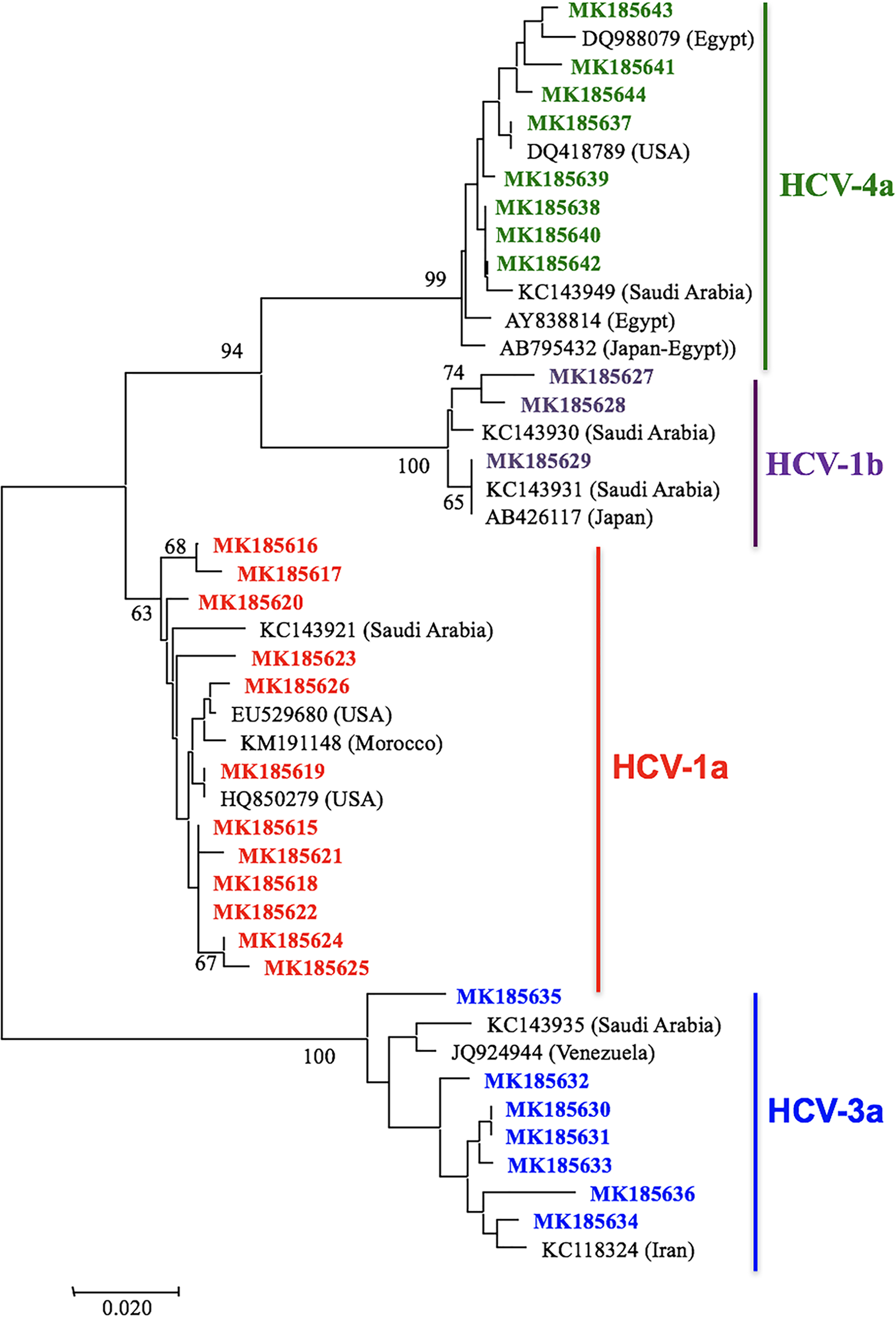
What are the benefits to using Lasergene for your analyses?
We do genetic epidemiology and genetic characterization of viral isolates, so we compare what we collect in Palestine with isolates from different parts of the world.
Since 2011, our research has centered on the viral genetics of viruses circulating among Palestinians. Lasergene has remained our primary analysis tool. My laboratory performed genotyping and mutation analysis of different viruses circulating in Palestine, Adenovirus (Qurei et al., 2012), HBV (Abdelnabi et al. 2014), influenza A (Bakri et al., 2019) and HCV (Rayan Da’as and Azzeh, 2019). One student has studied the genetic epidemiology of H. pylori (not yet published).
While our lab was originally learning to use MegAlign, I contacted Brian Anderson at DNASTAR several times for technical support issues or workflow questions and found him to be very helpful. On a few occasions, I was directed to Eric Ma for additional support. Later on, Brian Anderson was extremely generous and supportive, especially when I ran into a very difficult situation and needed to access Lasergene from a secondary location.
I briefly tried other options but stayed with DNASTAR because we found it more user-friendly than the alternatives. (N.B. See a selection of Maysa’s publications using MegAlign Pro below.)
Thank you, Dr. Azzeh, for letting us share you unique and exciting research on the DNASTAR blog!
If YOU are using DNASTAR software or services in your research and would like to be featured on our blog, please contact dnastar-info@dnastar.com.
Interested in comparing viral genomes or analyzing your own data?
Click the button below for a free trial of the complete Lasergene package, which includes MegAlign Pro!
Some of Dr. Azzeh’s publications citing MegAlign or MegAlign Pro
Rayan Da’as S, Azzeh M (2019) Subgenotyping and genetic variability of hepatitis C virus in Palestine. PLoS ONE 14(10): e0222799. https://doi.org/10.1371/journal.pone.0222799
Bakri M, Samuh M, Azzeh M (2019) Molecular epidemiology survey and characterization of human influenza A viruses circulating among Palestinians in East Jerusalem and the West Bank in 2015. PLoS ONE 14(3): e0213290. https://doi.org/10.1371/journal.pone.0213290
Abdelnabi Z, Saleh N, Baraghithi S, Glebe D, Azzeh M. “Subgenotypes and mutations in the S and polymerase genes of hepatitis B virus carriers in the West Bank, Palestine”. PLoS One. 2014:9(12). http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0113821
Qurei L, Seto D, Salah Z, Azzeh M (2012) A Molecular Epidemiology Survey of Respiratory Adenoviruses Circulating in Children Residing in Southern Palestine. PLoS ONE 7(8): e42732. https://doi.org/10.1371/journal.pone.0042732


Leave a Reply
Your email is safe with us.