For those working with gene panels or performing validation control workflows, BED and VCF files are integral to the entire process. In fact, through the Genome in a Bottle Consortium (GIAB), the National Institute of Standards and Technology (NIST) has developed a highly accurate and well-characterized set of genome-wide reference materials for NA12878, including BED and VCF files of high-quality sequence regions and variant calls, respectively. But what if you want to create your own custom BED and VCF files for use in your sequencing projects? By learning what BED and VCF files contain and how this information needs to be displayed, you can easily customize your own files.
VCF files contain information on your variants of interest. To successfully create a custom VCF file, you must set up your columns according to the specifications below:
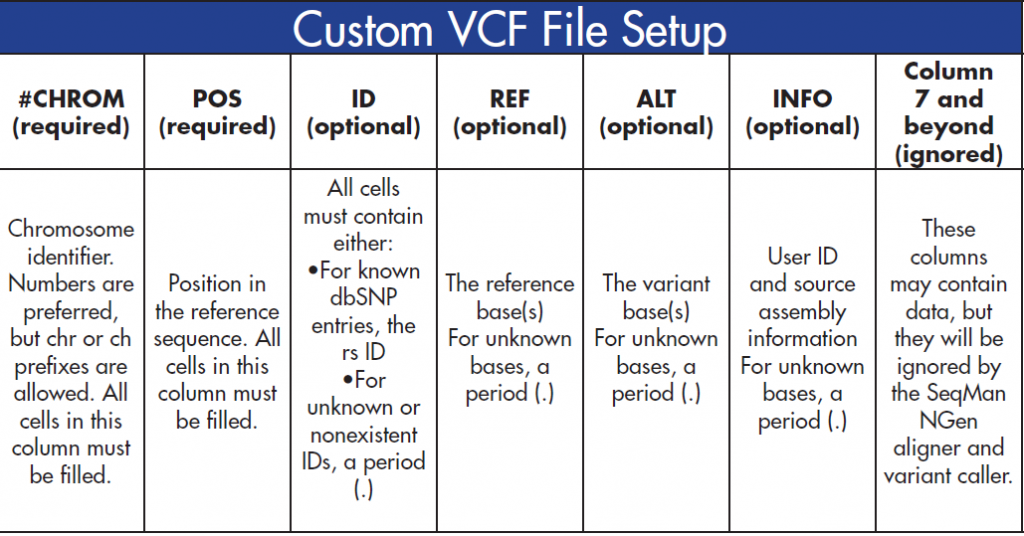
After your VCF file columns have been filled, it is important to sort the columns numerically, first by #CHROM and then by POS. Make sure to sort the columns numerically (1,2,3…) instead of alphabetically (1, 11, 12…)!
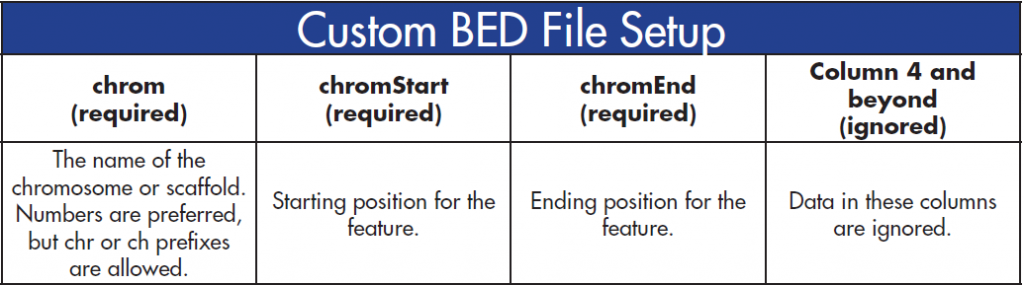
BED files are used to define capture regions in the assembly. The BED file column specifications are as follows:
Similar to the VCF files, the BED file columns must be sorted numerically by the first column and then by the second column. Additionally, all the cells in the first three columns must be filled.
Using these tips, you can easily create your own custom VCF and BED files for use in gene panel projects.



Leave a Reply
Your email is safe with us.