
Dr. Amanda Mitchell received her PhD in Biochemistry from the University of Wisconsin in Madison. Her thesis focus was on 3D structures of proteins from bacteria that modify sugars attached to antibiotics they produce. She joined DNASTAR in 2011 as a Quality Assurance Scientist, leading testing on Protean 3D and NovaFold. I recently sat down with Amanda to learn more about the function prediction capabilities in NovaFold.
What can you learn about a protein using NovaFold’s function prediction options?
NovaFold always predicts a model from protein sequence; function prediction is accomplished after the model has been predicted. When working with a protein of unknown structure, the function prediction step can help identify:
- Similar proteins that have already been solved;
- Potential ligand bindings of the protein;
- The function of the protein in the cell;
- What, if any, enzyme activity is associated with the protein.
How can researchers use this information?
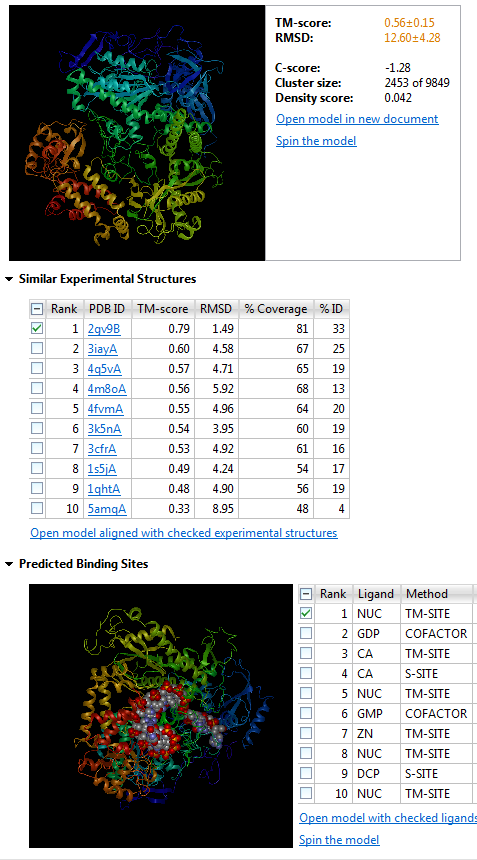
NovaFold results showing predicted model structure and binding sites for a Herpes 5 DNA polymerase sequence
There are a variety of applications, depending on the specific research objectives. For instance, I look at the similar experimental structures from a crystallography standpoint. Even after collecting the data for the structure, a scientist often does not know the shape of the protein. In order to solve the phasing problem of crystallography data collection, it is helpful to identify similar experimental structures. The proteins identified via function prediction can help you to identify potential proteins for molecular replacement, for example.
Additionally, looking at similar experimental structures might also highlight different functions associated with the predicted fold or a common function across all structures with a similar fold.
Predicted binding sites give you an idea of the protein’s active site(s). They may also suggest what additional molecules or atoms are required cofactors for an enzymatic reaction. For instance, in a DNA polymerase, you can get an idea of where the magnesium or calcium atoms might bind. Therefore, if you have an idea of what the function is or what the protein binds, and what residues are required for binding the substrate, you can infer where the protein active sites are. This can provide useful information if you are looking at modifying the protein via mutagenesis as it will allow you to potentially identify a handful of candidates that could be involved in substrate binding. Alternatively, if you are interested in drug targets, identifying the key residues involved would help you determine what interactions are required to allow your substrate of interest to bind.
The enzyme prediction feature gives you an idea of potential enzyme activity for a given protein. It can also corroborate the hypothesized protein function.
All of this information can help guide you in designing further experiments to corroborate the model and function predictions.
What might someone doing molecular biology or genomics work learn about a protein using NovaFold’s function prediction capability?
Everything from fold to function to potential active sites – basically just about everything you don’t know about a protein, and much more than you can learn from the sequence alone. Some examples of questions that can be addressed include the following:
- If you are looking at mRNA sequences and want to know what proteins are being produced – and are those proteins wild type or not?
- If the proteins aren’t wild type, what implications does that have?
- Do the mutations cause a shift in the protein’s function?
- Does it destroy the active site?
- What is the effect of mutation on protein?
A predicted model with function prediction from NovaFold could answer all of these questions and many more.
What advantages does NovaFold provide over traditional methods of inferring protein function?
Traditionally users would have to search the PDB extensively, or use open-source tools to get protein function information. Then they would need to piece together all of the information manually. This often amounts to days or weeks of work. By contrast, NovaFold automatically shows all the predicted models and function information in one document and allows you to save and quickly reopen results.
How can someone use NovaFold to predict protein function based off a protein sequence?
Users just need to submit a sequence to NovaFold without modifying default parameters – function prediction is on by default. Typically results will include five models. NovaFold runs function prediction on the top model plus all additional models with a confidence score of 0.5 or higher, giving the user more confidence in the function prediction results.
Interested in trying NovaFold for protein structure and function prediction? Request a quote here!



Leave a Reply
Your email is safe with us.