We recently presented a webinar entitled “Mastering Phylogenetic Tree Creation & Optimization with MegAlign Pro.” You can view the recording of this webinar using the link. Other archived webinars can be found on our Webinars page.
Due to the enthusiastic participation of our record audience, presenter Dr. Brian Walsh did not have sufficient time to answer everyone’s questions. As with other recent webinars, we therefore decided to turn a selection of your questions into the following “Q & A” blog post.
Topics Covered in this Article
General MegAlign Pro Questions
- How can I get this software?
Do any of the following: Request a 14-day free trial / Purchase online / Request a quote
- What advantage does MegAlign Pro have compared to PAUP*?
Like MegAlign Pro, PAUP* is a powerful program. However, MegAlign Pro has two main advantages:
- MegAlign Pro can be used to create the alignment as well as to analyze it. PAUP* can open alignments but does not generate them. The alignment step would require the use of additional software.
- MegAlign Pro has a much better-developed user interface, and most people find it much more intuitive to use.
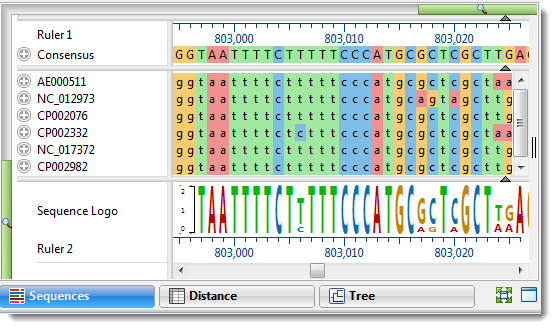
Performing a Multiple Alignment
- Which multiple alignment method is best for different situations? …and… Which methods are most appropriate for aligning gene, CDS, and protein sequences?
All four of MegAlign Pro’s gene-level alignment methods (Clustal W, MUSCLE, Clustal Omega and MAFFT) work with any type of sequence. However, depending on the data being used, these methods will all involve some tradeoffs between speed, accuracy, and customization. Other things to take into account when choosing a method include the lengths of the sequences, number of taxa involved, and their relatedness. For an easy-to-understand description of when to use each method, see our recent blog post, Two Ways to Find the Best MegAlign Pro Multiple Sequence Alignment Method.
- Is it more appropriate to deduce phylogenetic relatedness using gene sequence, CDS sequence, or something else?
See Brian answer this question. In short, this varies depending on the problem you are trying to solve. For instance, Brian studied very closely related pepper plant species, so he chose to compare intron sequences, since there was more variability in these non-coding regions. If he had instead aligned gene or CDS sequences, they would have been too similar to allow him to build an accurate phylogenetic tree. Similarly, Brian believes the non-coding region he was using was too divergent to analyze across subfamilies of Solanaceae.
- What’s the difference between Mauve and MAAFT?
Mauve is for genome-level alignments while MAFFT is one of four methods available in MegAlign Pro for gene-level alignments. For descriptions of each alignment method, see the User Guide topic Multiple alignment methods and options. Additional information on choosing a method can be found in our recent blog post Two Ways to Find the Best MegAlign Pro Multiple Sequence Alignment Method.
- Is it possible to restrict mRNA alignments to only the CDS of a transcript? Similarly, is it possible to limit a gene alignment to CDS, and CDS alignment to Protein?
A great way to approach this would be to edit your sequences in SeqBuilder Pro in order to generate new sequences that consist only of the CDS region or other region of interest. It is even possible to concatenate sequence from different genes.
- For huge sequences, could I align portions of them at first, then integrate the subtrees into one big tree? is this logically correct and possible?
It’s not clear whether you’re referring to the number or length of the sequences, but the answer is the same either way. Phylogenetic analyses are approximations of evolutionary history based on the data provided. Changes to a dataset will change the phylogenetic results: maybe a little, maybe a lot. These analyses can be compared but not combined.
If you have a large number of samples, you can analyze a pared-down sampling of a broad group. For instance, you could include 2-3 taxa from each genus within a family-level analysis. Before doing so, we recommend running many analyses on subsets of the data, so you have a good idea of the relative relationships among your samples. In the end, you can present the overall ‘pared-down’ tree as well as separate smaller trees that flesh out the full range of the clades/relationships.
Handling gaps after the initial alignment
- Is it better to remove gaps by trimming out the gapped areas prior to (re)aligning, or to remove gaps later on by using a pairwise alignment?
After you initially align your sequences, gapped regions appear in gray in MegAlign Pro’s Overview.
When publishing, you need to report precisely what was excised and why. So, we do not recommend removing small indels, as they have a minor effect on the quality of phylogenetic analysis. We would limit trimming to data at the ends of the alignment and to very long indels that do not contain useful information.
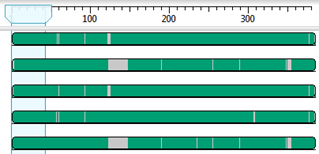
If there are large gaps in some sequences, it’s best to remove that portion through trimming and then to perform a realignment. Some situations that can cause these large gaps include 1) sequences of very different lengths such that some sequences will have no data on the 3’ and/or 5’ ends; and 2) a few sequences containing a long INDEL that is not present in other sequences, and therefore appears as gaps for those where the INDEL is not present.
For detailed instructions on how to trim sequences, see the MegAlign Pro User Guide topics trimming the ends from an alignment and trimming an individual sequence. See Brian demonstrate how to remove large gaps on either end prior to realigning.
- For amino acid sequences that have gaps relative to each other when you align them using one method (e.g., Clustal Omega), would you recommend realigning using a different method (e.g., MAFFT)?
We definitely recommend trying different alignment methods with each data set. There is no hard and fast rule about which method will work with a given set, so it’s best to try several. Often, you will get the same results for each method, but in some cases, one method may be a clear “winner” for your data set.
- How do you remove a large gap from the middle?
MegAlign Pro does not support removing large gaps from the middle of an alignment at this time.
Distance Table
- In the Distance table, which sequences are more closely related: those with a low value, or those with a high value?
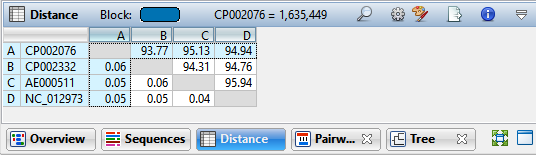
It depends what metric you are looking at. If you are viewing “uncorrected pairwise distance,” the distance becomes smaller as the relationship becomes closer. If you are looking at “% similar” or “% identity,” it is the opposite; a larger number indicates a closer relationship.
- Can you sort percent identity of a single reference sequence to all the other sequences in your alignment?
MegAlign Pro does not currently do this, but this is a fantastic idea. In fact, Brian just submitted this as an enhancement request. Thank you! We are always looking for ways to improve our software.
Building Phylogenetic Trees
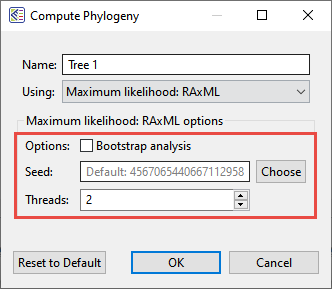
- How important are bootstrap values?
MegAlign Pro lets you select bootstrap analysis options when computing a Maximum likelihood: RaxML tree (see image). See Brian answer this question.
- Which method should I use for building my phylogenetic tree?
Brian recently covered this topic in detail in his blog post How to Create the Best Phylogenetic Tree for Your Data Using MegAlign Pro. You can also see Brian answer this question during the webinar. The short answer is that Brian actually uses all of these methods. For instance, he may apply them to the same data set and compare similarities and differences of the results within a publication. Many times, all the methods will yield essentially the same results.
- How do I reroot trees?
Use the Tree > Root On command. This is explained in the User Guide topic Reroot a tree.
- I need to have the organism name in italics. How can I do this in MegAlign Pro?
Simply choose View > Style > Tree and press the button with the letter “I.” See Brian demonstrate this. This and other Tree section options are described in the User Guide topic Tree Section.
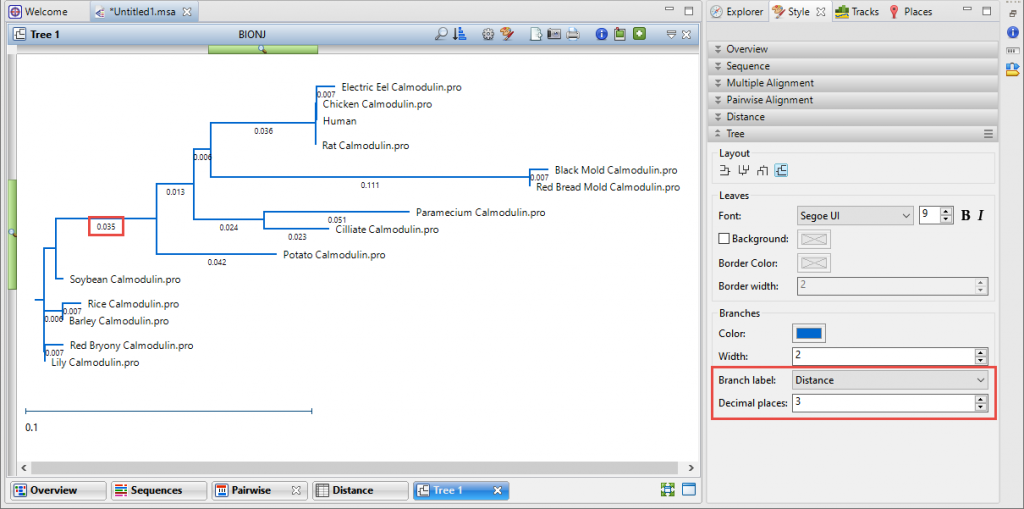
- When looking at the tree, where do I see the distance numbers? How can I understand the scale?
By default, the “uncorrected pairwise distance” numbers are displayed under the tree branches. Smaller distance values indicate closer genetic relationships. If you don’t see a number under the branches, or if you want to show a different number of decimal places, choose View > Style > Tree and make changes using the Branch label menu and Decimal places field. The Tree section options are explained in detail in the User Guide topic Tree Section.
- I used to believe that Maximum Likelihood methods were computationally more complex than distances methods. In your example in the webinar, Maximum Likelihood was faster than Neighbor Joining. Was that due to the use of multiple threads?
Our RaxML example in the webinar used just two threads, which could have played a role in this algorithm completing sooner. However, RaxML is also specifically optimized to analyze thousands of samples quickly.
- Is there a way to create a circular tree design?
Unfortunately, MegAlign Pro is not currently able to create a circular tree, but it is in our list of features we are working toward implementing. For now, you can accomplish this by exporting your tree file for use in a tree visualization program such as FigTree. See this User Guide topic to learn how to export a tree from MegAlign Pro.
- Is there any option for Bayesian method? How about evolutionary model testing?
MegAlign Pro does not currently include Bayesian Inference analyses, but this is high on our list of planned enhancements. We developed evolutionary model testing, but it has not yet been implemented because RaxML is limited to the GTR+G+I model. Testing for statistical overfitting of a model does not fit with the phylogenetic options at this time.
- Any tips for making a tree for viruses, especially COVID-19 genomes? How about for fungi?
In reality, most of the alignment algorithms researchers rely on were not designed for working with genomes, even small ones. So long data sets can really push the alignment algorithms to their limits. We recommend using the MAFFT algorithm with a few custom settings. From the Align drop menu choose Align with Options. Change the Algorithm to Very Fast, Progressive. You may furthermore need to remove some of the sequences and try the alignment again until the alignment is successful. During our in-house tests, for example, 2000 COVID-19 genomes (~30kb) would successfully align, but 3000 would not.
- Sometimes MegAlign Pro fails to generate a Tree and shows only a red outline. What are reasons that this might happen, and how would I prevent it?
We have seen this ourselves when working with many very divergent sequences/samples, especially divergent protein sequences. In this case, the degree of highly divergent data is beyond what the alignment algorithms can handle. When this happens, the phylogenetic tree is fully collapsed into a red box, and the distance table is populated with NA instead of numerical values.
Our developers are investigating this, but I’m afraid we do not have a great solution to offer. In the case of divergent protein sequences, you could try using the pre- translation nucleotide data, as this is likely to be more conserved.
Exporting Trees
- Although great, FigTree has not been updated since 2018 (note: this is still true as of Dec 2023). What software would you recommend using to generate more sophisticated Tree displays using a Mac?
See Brian answer this question. After the webinar, we did some research and found the following open source alternatives that all work on Mac: TreeView X, Archaeopteryx and Dendroscope. We have not used any of these ourselves so cannot offer advice on the pros and cons of each.
- How can trees be shared with colleagues that may not have access to the software, i.e., image export, screen shot, html?
See Brian answer this question. All of MegAlign Pro’s image and data export options are listed in the User Guide topic Export. Click any of the links for detailed information.


Leave a Reply
Your email is safe with us.