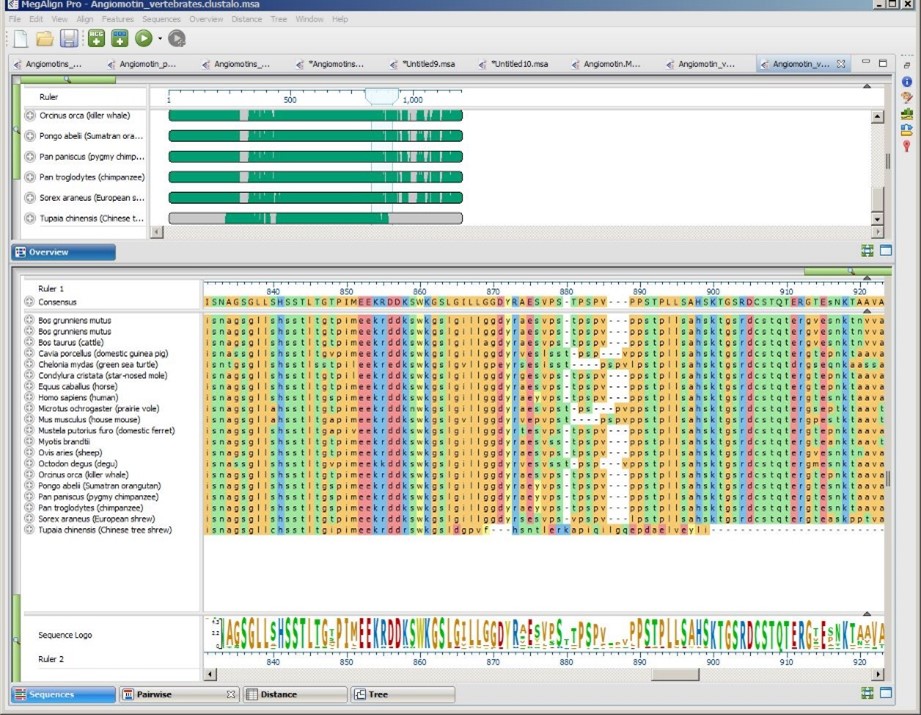
If your work involves the study of evolutionary relationships, you know that it can be hard to choose the right multiple sequence alignment (MSA) algorithm for your data.
If you’re doing a whole-genome alignment in MegAlign Pro and have nucleotide sequences, there’s no contest: Mauve is the best MegAlign Pro algorithm for you. But what if you are doing the more common gene-level alignments? MegAlign Pro offers four popular algorithms that work for both nucleotide and protein sequences: Clustal Omega, Clustal W, MAFFT and MUSCLE. These algorithms all have user-editable options for speed, capacity, algorithm and more. Which of these methods is the “best”?
A published comparison of the same four gene-level alignment algorithms available in MegAlign Pro showed that no one method was universally superior to the others. Algorithms that worked great with one data set could be the worst option for a different data set. That’s why it’s important to try different alignment algorithms and settings to learn which ones produce the best result for your data.
That said, it is possible to make general recommendations about the best starting algorithm for a particular situation. In this blog post, we’ll provide two solutions for choosing the best “starting” alignment algorithm for your needs.
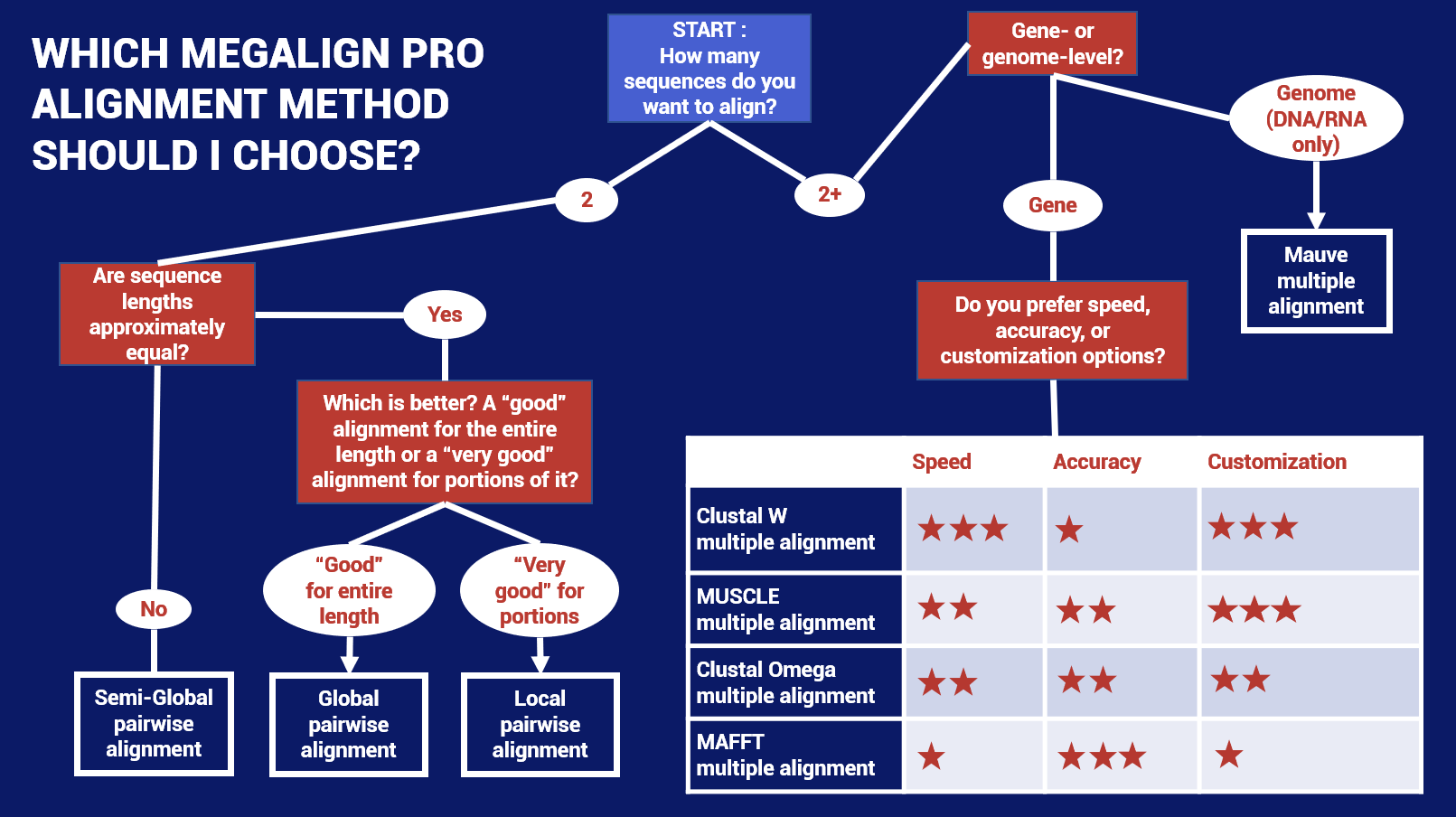
Option 1: Choose based on speed, accuracy and/or customization options
If you want to choose the perfect balance between speed, accuracy and/or customization, we have developed the following flowchart to help you choose the best starting algorithm. Note that this chart includes MegAlign Pro’s multiple and pairwise alignment methods.
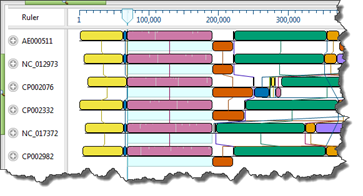
Option 2: Optimize for capacity or special circumstances
If you prefer to optimize based on the number and/or size of your sequences or on other criteria, choose your situation from the following list to see which method we recommend that you start with.
I have genome-length nucleotide sequences OR…
My nucleotide sequences are not on the same strand OR…
My nucleotide sequences contain large rearrangements (e.g., inversions, translocations)
Use Mauve. Mauve is MegAlign Pro’s only genome-level aligner and only algorithm capable of producing a multi-block alignment or an alignment when one or more of the sequences are rearranged relative to one another. Mauve uses MUSCLE to create multiple alignments for each block that contains more than a single sequence. The main disadvantage with Mauve is that its fine-scale gapping not as good as MegAlign Pro’s four gene-level alignment methods described below.
Mauve was originally developed by Aaron Darling, Bob Mau, and Nicole Perna in 2010 at the Genome Evolution Laboratory at the University of Wisconsin-Madison.
Links:
- MegAlign Pro tutorial “Genomic alignment with Mauve”
- MAUVE website
I have fewer than fifty “short” (< 1kb) DNA, RNA, or protein sequences
Start with ClustalW. The Clustal W alignment algorithm is faster than Clustal Omega (reference), though its maximum accuracy is only obtained if you select the default “Slow-Accurate” option in MegAlign Pro’s ClustalW settings panel. A disadvantage to this method is that it does not always handle end gaps ideally. ClustalW was developed by JD Thompson et al. in 1994 at the European Molecular Biology Laboratory, Heidelberg, Germany.
Links:
I need to do a gene-level alignment of up to thousands of DNA, RNA, or protein sequences AND/OR…
I want to specify that one of the sequences be used as a reference sequence for the alignment
Start with MAFFT. The MAFFT alignment algorithm is based on Fourier transformation and has several editable options. At least one paper found it to be the most accurate of the four gene-level algorithms (reference). Another found that it gave “structurally consistent alignments” for RNA data specifically (reference). When using long sequences, the algorithm performs best if the sequences are closely related. MAFFT was developed by Katoh M & Kumar M (2002) at the Computational Biology Research Center.
UPDATE (2/22/22): With the release of Lasergene 17.3.1 in January 2022, MAFFT now supports alignment of up to 10,000 viral genome sequences. The updated MAFFT algorithm also allows you to specify a reference sequence. To learn how to do this, see the MegAlign Pro User Guide topic MAFFT alignment options.
Links:
I need to do a gene-level alignment of up to thousands of taxa and have DNA, RNA, or protein sequences
Start with MUSCLE. The MUSCLE sequence alignment method has many editable options and one paper found it to be faster than Clustal Omega alignment (reference).
MUSCLE was developed by independent bioinformatician Dr. Robert Edgar in 2004.
Links:
- MegAlign Pro tutorial “MUSCLE alignment with multi-segment sequences”
- MUSCLE website
- MUSCLE manual
- Article describing the algorithm
I have a small number of protein sequences
Start with Clustal Omega. This method was designed for protein sequences but can also be used for nucleotides. It has several editable options. The developers state it is more accurate than ClustalW. It’s also very fast, aligning hundreds of thousands of sequences in a few hours. Clustal Omega was developed by F Sievers et al. in 2011 at University College Dublin.
UPDATE (2/22/22): If you have a large number of protein (or any other type of sequences), use MAFFT rather than Clustal Omega. The version of MAFFT included in Lasergene 17.3.1 (released Jan. 2022) has the highest capacity of any available alignment algorithm.
Links:
We hope this blog post has given you some ideas for how to choose a multiple alignment method that will work as the optimal starting point for your data set.
Would you like to try these workflows for yourself? Click the button to request a fully-functional 14-day free trial of Lasergene, including the MegAlign Pro application. Both downloadable and online trials are available.
Want to learn more or see a video about multiple sequence alignment?
Click the button to visit our “Multiple Sequence Alignment” workflow page.



Leave a Reply
Your email is safe with us.