The Multiple alignment and Pairwise alignment sections allow you to select color schemes for sequences in the Sequences and Pairwise views, respectively. Each available color scheme is described below:
| Color Scheme | Description | Nucleotide Legend | Amino Acid Legend |
|---|---|---|---|
| Color by Chemistry | The default color scheme used by multiple DNASTAR applications. Amino acids are colored according to their side chain chemistry. While Color by Chemistry can be used for nucleotide sequences too, we recommend instead using a Solid background color for nucleotides and then selecting a Comparison like Color only non-conserved residues, Color only differences from reference, etc. |  |
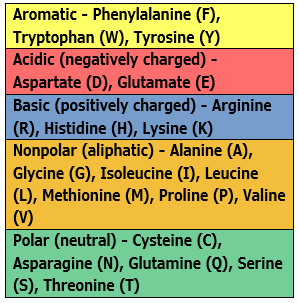 |
| Physiochemical: Shapely | Matches the RasMol amino acid and RasMol nucleotide color schemes, which are, in turn, based on Robert Fletterick’s “Shapely models.” |  |
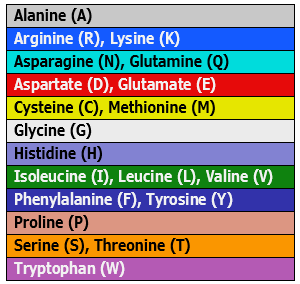 |
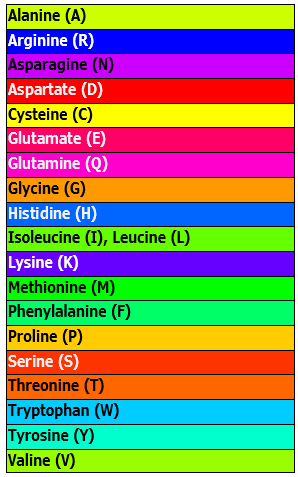
| Physiochemical: Zappo | Colors residues according to their physico-chemical properties, and is also used in JalView (Waterhouse et al., 2009). |  |
 |
| Physiochemical: UGene | Unipro UGENE coloring scheme. |  |
 |
| Spectral Colors: Taylor | Taken from Taylor (1997) and also used in JalView (Waterhouse et al., 2009). |  |
 |
| Clustal X | Taken from Thompson et al. (1994). This color scheme is only available for protein alignments. | N/A |  |
| Grayscale – Percentage Identity | This option uses only black/white/gray and is suitable for publications requiring grayscale images. |  |
 |
| Percentage Identity | This option uses only shades of slate blue. |  |
 |
Need more help with this?
Contact DNASTAR


