To export the alignment report for the selected block from a Mauve alignment, use File > Export Data > Alignment Report. To export the report for any other alignment type, use File > Export Data > Alignment Report. In both cases, the report is saved as a .txt file.
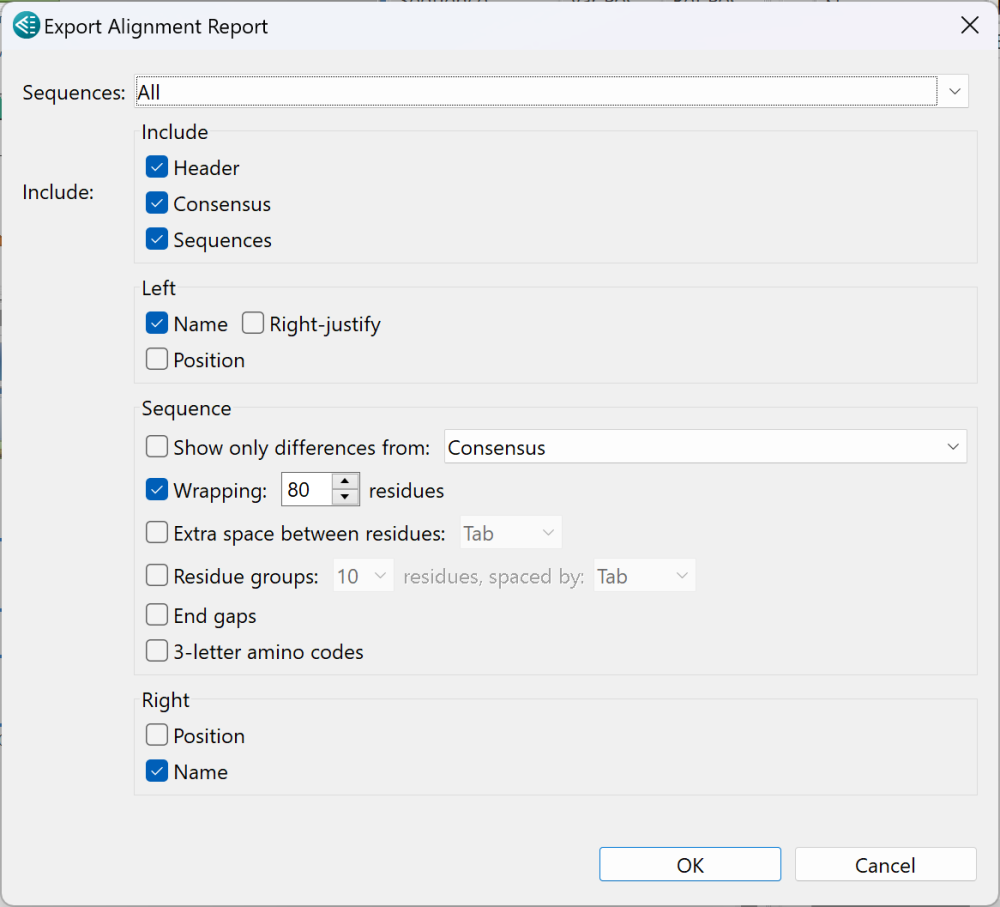
- From the Sequences drop-down menu, choose All to create a report for all sequences or Top to create a report only for the uppermost sequence in the Sequences view.
- In the Include section, check items you with to include and remove checkmarks from the others.
- In the Left and Right sections, check boxes showing where, if anywhere, you wish to include the sequence Name and/or Position. To right-justify any labels chosen to occupy the left margin, check Right-justify.
- To include only residues that differ from a baseline or comparison sequence, check Show only differences from and choose the sequence from the drop-down menu. Options include the Consensus sequence or any of the sequences in the project.
- To wrap sequences (rather than allowing them to go off the page), check Wrapping to wrap sequences in the report rather than allowing the sequence to go off the page. If you elect to wrap, enter the number of residues per line in the text box.
- To include a tab or space between residues, check the Extra space between residues tab and choose Tab or Space from the drop-down menu.
- To group residues and include a tab or space between them, check Residue groups, make a numberican selection from the drop-down menu, then choose Tab or Space from the second drop-down menu.
- To include gaps at the end of the sequence, check End gaps.
- If your alignment consists of protein sequences, you can show their 3-letter IUPAC amino acid codes in the report by checking 3-letter amino codes.
After making the desired selections, press OK to generate the report. You will then be prompted to save the report.
Need more help with this?
Contact DNASTAR


