The Reference Sequence wizard screen is the first of five screens used to set up a gene homology alignment. You must input at least one reference sequence here before proceeding.
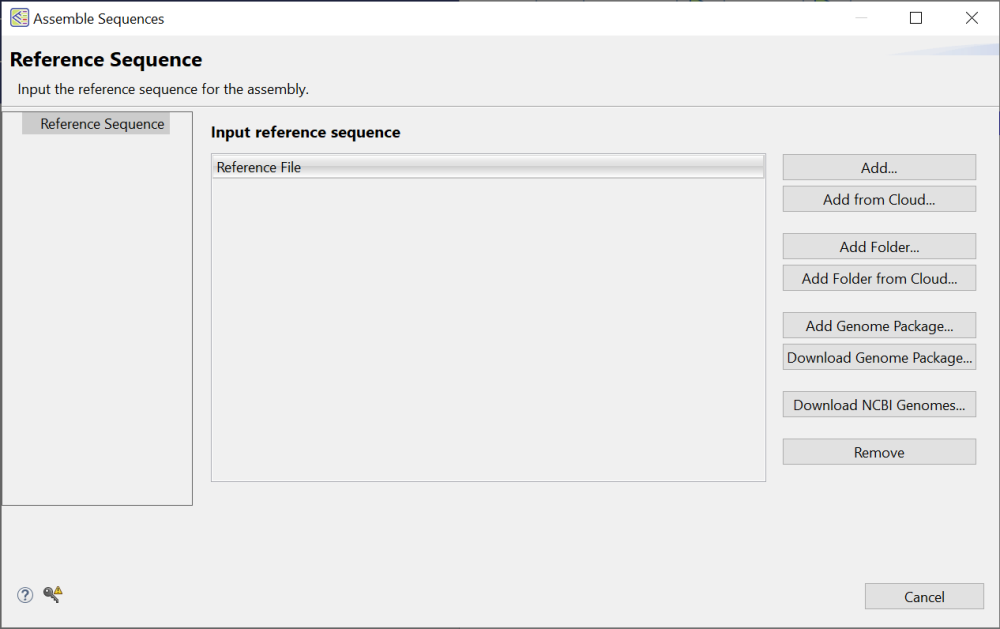
To add one or more sequences from your computer:
To add local sequences, press Add or Add Folder. Navigate to and select the desired file(s) or folder of files, then click Open.
To add one or more sequences from the DNASTAR Cloud Data Drive:
Press Add from Cloud or Add Folder from Cloud. Navigate to and select the desired file(s) or folder of files, then click the green check mark (
The first time you press a button associated with the Cloud Data Drive, you may be prompted to enter your Email and Password. Enter the information that you use when logging in to the DNASTAR website. Check the Save password box if you would like MegAlign Pro to save this information so you do not need to retype it in the future. If you are a licensed Cloud user, a message will display the number of assemblies remaining on your license. Click OK to close the message and continue to the next screen. If you are not licensed, a different message will appear, asking you to purchase a Cloud license through DNASTAR Support (support@dnastar.com).
To add a genome template package for a common model organism:
To add a curated genome template package from DNASTAR, press Download Genome Package and select a package from the list.
If you previously downloaded the same package, you can skip this step and instead press Add Genome Package. In this case, navigate to the package location and click Open. Note that the Add Genome Package and Download Genome Package buttons are disabled if you have already added files using the Add or Add Folder buttons.
To add a genome directly from the NCBI database:
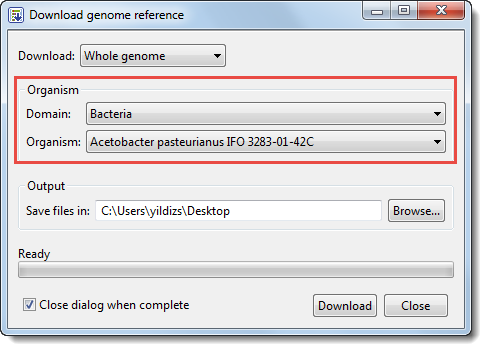
- Press the Download NCBI Genomes button. This launches the Download genome reference dialog.
- Use the Download dropdown menu to choose between downloading a Whole genome or By accession numbers.
- If you select Whole genome, MegAlign Pro will retrieve the most recent build of the selected genome. Use the next two drop-down menus to select the Organism type and Organism.
MegAlign Pro will download all the reference sequences from the NCBI Entrez Genome Project database for the selected genome. These downloads may include auxiliary genomes such as mitochondria and chloroplasts. They may also include some contigs which have not yet been placed by the genome finishing process.
- If you select By accession numbers, the Organism section disappears and is replaced by an Accession numbers entry area. Type an accession number or paste it from your clipboard, then press Add to add a number to the list. You can type accession numbers with or without explicit version numbers (e.g., NC_000913.3 or NC_000913). If the version number is omitted, then NCBI’s latest version is returned.
Continue adding numbers, as desired. Multiple accession numbers should be separated using a space, comma, semi-colon or line break. To remove an accession number from the list, select it and click Remove.
- If you select Whole genome, MegAlign Pro will retrieve the most recent build of the selected genome. Use the next two drop-down menus to select the Organism type and Organism.
- Click Browse to select a name and location in which to save the downloaded genome files.
- If you do not need to download additional genomes, you can check Close dialog when complete. Otherwise, leave the box unchecked to keep the dialog open after initiating the current download.
- Press Download. Once the download is complete, a message like the one below will appear.
- Click OK to close the dialog and add the accessions to the Reference Sequence screen. If you checked Close dialog when complete, the Download genome reference dialog will also close. Otherwise, it will remain open so that you can download additional NCBI genomes.
____________________________________________________________
Regardless of the method used to add sequences, you can remove a sequence from the list by selecting it on the left and then pressing the Remove button on the right.
Once the reference sequence(s) have been added, click Next to proceed to the Input Sequences screen.
Need more help with this?
Contact DNASTAR