To select which columns to display in the Variants view, or to rename or reorder them, click the Choose or rearrange columns (
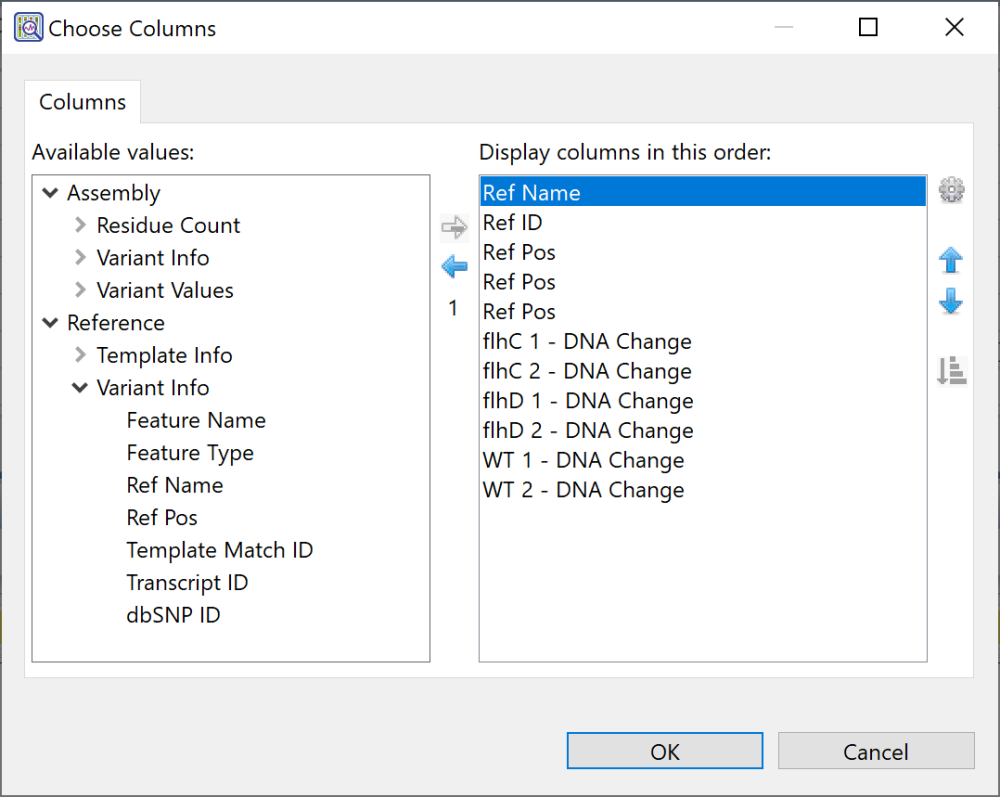
Available but yet-unused columns are on the left, while currently-applied columns are on the right.
The available options and their parent categories depend on whether a single-sample or multi-sample version of the Variants view is active. Phased genome assemblies and human assemblies that included annotations from the Variant Annotation Database (i.e., by checking a box in the SeqMan NGen wizard) will have additional options compared to other types of assemblies.
Using the Choose Columns dialog:
| Task | How To |
|---|---|
| Expand a category to see available sub-options | Click the gray arrow next to the category name. Some sub-options can be expanded further to show another level of options. For example, if the Variants view includes multiple samples, you can expand many of the column options and elect to apply data for only one of the samples to the table. |
| Add a column to the display | Select its name on the left and press the right arrow key to move it to the right. Selecting a parent category selects all sub-options within that category. |
| Remove a column from the display | Select its name on the right and press the left arrow key to move it to the left. |
| Change the order of displayed columns | Select the column name you wish to move on the right, then use the up/down arrows ( |
| Sort selected columns alphabetically | Choose columns in the right pane using Shift+click or Ctrl/Cmd+click. Then click the Sort selected columns by name tool ( |
| Change the name of a column | Select its name on the right, then choose the Configure column tool ( ) from within the Choose Columns dialog (i.e., not the tool with the same icon in the Variants view header). In the popup dialog, type in the desired name and press OK. ) from within the Choose Columns dialog (i.e., not the tool with the same icon in the Variants view header). In the popup dialog, type in the desired name and press OK.  |
| Select display options | If a single-sample Variants view is active, the Choose Columns dialog has two tabs: Columns and Options. Click the Options tab and check or uncheck boxes to specify whether you wish to Show (residue) counts as percents or Show codon bases and distance to feature. 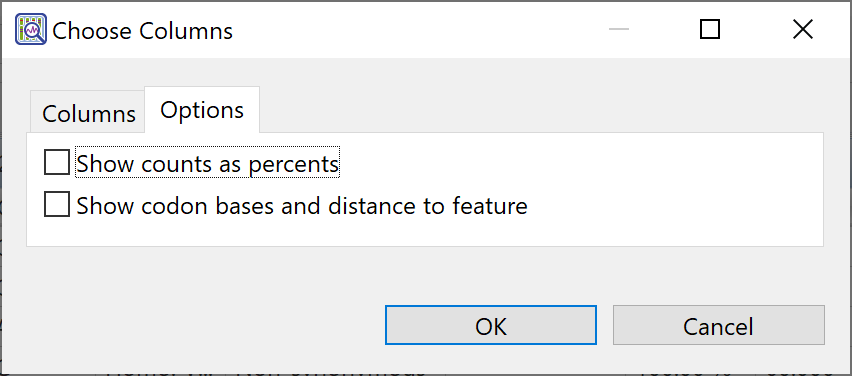 Note: Another way to see residue counts as a percent is to use the Show counts as percent tool (  ) in the Variants view header. ) in the Variants view header. |
Columns that can be applied to the Variants view are listed below in alphabetical order:
| Column Type | Description |
|---|---|
| Amino Acid Change | This column is only available if Show Codon Bases & Distance to feature is unchecked. The column shows the change(s) in the amino acid sequence, using the nomenclature established by the Human Genome Variation Society and The Sequence Ontology Project. This includes:
|
| AnnoID | (DNASTAR internal use only). |
| Called Base | The dominant variant in the aligned column. In the case of a heterozygote call, both bases at the position are shown, separated by a vertical bar. For multi-base insertions, the inserted string is shown. For multi-base deletions, the deleted bases are represented with dashes (-). |
| Coding Feature Distance | Shows whether variants are within or near a named feature, and the distance from that feature. For .assembly files and certain .sqd files (e.g., from de novo or special templated workflows), the following color scheme may be used:
|
| Codon | When a translated feature is present on the reference sequence at the position of a variant, a codon change is displayed. The codon and amino acid translation is shown for the reference sequence and compared to the codon and amino acid translation for the selected variant. The position number of the amino acid change is also displayed. |
| Cons Pos / Contig Pos | The position on a gapped contig corresponding to this chromosome (for SeqMan Ultra and SeqMan NGen assemblies only). |
| COSMIC | The Catalogue of Somatic Mutations in Cancer (COSMIC) ID for positions with known variants. Double-clicking on the entry opens the corresponding page at COSMIC. For human assemblies only. |
| Cross Compare / cross | (DNASTAR internal use only) |
| dbSNP ID | The dbSNP rs ID, if available, for positions with known variants. Double-clicking on the entry opens the corresponding page at dbSNP. |
| Deletion | The number of deleted bases in the Indel. |
| Depth | The number of reads overlapping the aligned column. Since this calculation disregards bases below the quality threshold, the Alignment View may show a greater number of sequences than the Depth shown in the Variants Summary Report. The default quality threshold for assembly in SeqMan NGen is 5. |
| DNA Change | Change(s) in the DNA sequence affecting either CDS features or splice sites are indicated using the nomenclature established by the Human Genome Variation Society (HGVS). A “c.” prefix, followed by coordinates taken from the ORF, denotes a change in a CDS feature. For example:
|
| Feature Name | If a variant is located within an annotated feature in the reference sequence, the feature type and name are displayed. A single nucleotide change may sometimes be reported as affecting multiple overlapping features. These can include different overlapping genes on the same or opposite strands, as well as alternatively spliced messages from the same gene. In this case, SeqMan NGen produces multiple VCF Variant table entries at the same position, one for each reported feature. A bracketed number follows the Feature Name to indicate which isoform from the Feature view table was used (e.g., TP53 [2]). Note: If a non-gene feature (“mRNA”, “CDS”, etc.) exists in the template file, but has no corresponding “gene” feature, SeqMan NGen adds the “gene” feature automatically during assembly. The locations of any automatically added “gene” annotations are indicated by asterisks (*) in this column. |
| Feature Type | For variants within a gene feature, the feature type is shown in the following order of precedence: CDS, mRNA, Gene. If Show Codon Bases Distance to feature is selected, this column also contains a feature designation if the variant is within 150 bases of the nearest exon. Therefore, it is possible for a variant that is in a gene to also be listed as a CDS, mRNA, etc. When Show Codon Bases is checked, the Feature Type column will also show the distance to the nearby exon and an arrow indicating the direction of the feature. Feature types for different variant locations are shown below:
|
| Genotype | When the “Diploid” SNP detection method is used in a SeqMan NGen assembly, there are four possibilities: 1) homozygous variant (both alleles have the same base and it is different from the reference), 2) reference (both alleles have the same base and it is the same as the reference), 3) heterozygous reference (two different alleles are called, one with the same base as the reference, the other with a variant base), and 4) heterozygous not reference (two different alleles, neither of which match the reference base). It is quite rare for the reference case to occur in the table. This only happens in cases where there is sufficient evidence of the possibility of a variant to pass the filtering threshold, but where the evidence is still quite weak. These cases are usually eliminated by even modest filtering. When the Haploid SNP detection method is used, only variant and reference are possible. Note: In this column, if one or more of the adjacent variants is called as a heterozygote, the coalesced variant is also called a heterozygote. Therefore, for a coalesced variant to be called homozygous, all positions must be called homozygous. |
| GERP | The Genomic Evolutionary Rate Profiling (GERP) score representing the calculated evolutionary constraint at that position. GERP data is automatically delivered when you use DNASTAR’s human template package prior to performing a templated assembly in SeqMan NGen. To limit the size of the data file required, only positions with scores of 1.0 or greater are displayed. GERP is a tool that provides a score for each position in the human genome that estimates whether that position is under purifying selection or not (Davydov et al. 2010). GERP uses alignments between the human genome and 33 other mammalian genomes to quantitate the position-specific constraint in terms of rejected substitutions, defined as the difference between the neutral rate of substitution and the observed rate, estimated by maximum likelihood. Substitutions in sites under selection are assumed to be more deleterious than those not under selection. Scores range from negative values to ~6. Positions with scores below or near zero are not under selection. Conversely, the more positive the score, the more constrained the position. GERP information can be useful in evaluating the impact of non-synonymous variants in coding regions and the impact of changes in or near promoter elements, among others. |
| Homopolymer | Indicates whether the variant occurs within a homopolymeric run, which is defined as two or more identical bases in a row. When using Pacific Biosciences (PacBio) or Ion Torrent data, SeqMan Pro may not list all homopolymeric indels. When possible, insertions or deletions are placed at the 5’ end (top strand) of the run during alignment. |
| Impact | The impact of the variant or indel on the genome, displayed as one of the following values:
|
| Is Phased | Displays the word “phased” if the variant is in a phase block. Phase blocks are regions of the reference that could be phased into two haplotypes. |
| Length | Length of the variant in bases. |
| Long Ref Name | (DNASTAR internal use only). |
| MM Count 1 | Count of articles in the Mastermind Genomic Search Engine with cDNA matches for this specific variant. See View Mastermind Genomic Search Engine results online. |
| MM Count 2 | Count of articles in the Mastermind Genomic Search Engine with variants either explicitly matching at the cDNA level or given only at protein level. See View Mastermind Genomic Search Engine results online. |
| MM Count 3 | Count of Mastermind articles in the Mastermind Genomic Search Engine, including other DNA-level variants resulting in the same amino acid change. See View Mastermind Genomic Search Engine results online. |
| MM Gene | Genes for this variant from the Mastermind Genomic Search Engine. See View Mastermind Genomic Search Engine results online. |
| MM HGVS | HGVS genomic notation for this variant from the Mastermind Genomic Search Engine. See View Mastermind Genomic Search Engine results online. |
| MM ID 3 | Variant identifiers in the Mastermind Genomic Search Engine, as gene:key, for MMCNT3. See View Mastermind Genomic Search Engine results online. |
| MM URI 3 | Search URI for articles in the Mastermind Genomic Search Engine, including other DNA-level variants resulting in the same amino acid change. See View Mastermind Genomic Search Engine results online. |
| PDB ID | Worldwide Protein Data Bank (PDB) ID number. |
| Phase Block | Either displays the word “unphased” or the number of the phase block. Phase blocks are regions of the reference that can be phased into two haplotypes. Unphased reads either lack heterozygous sites or the pattern of these sites does not match the other reads. |
| Phase Consistency | A quality or confidence score for the phasing calls. GenVision Pro’s phasing algorithm tends to pick the longest span with the most reads. This usually results in accurate phasing results. However, multiple competing patterns like structural variations and repeats can confuse the assignment of a read to a particular A/B allele. This means that GenVision Pro can sometimes pick sites based upon alignment artifacts rather than “true” heterozygous sites. The Phase Consistency score helps determine the trustworthiness of phasing results at each location. |
| Phased Bases | The base call for each allele falling within a phase block. If both alleles have the same base, only a single base is shown in this column. |
| P Not ref | The probability that this position does not match the reference. For combined SNPs and indels, P not ref will be the minimum of the P not refs in the used columns. |
| Q call | The Phred-like quality score of the called genotype. It is a measure of the confidence that the SNP is present in the sample on a 0-60 log10 scale. For combined SNPs and indels, Q call will be the minimum of all available columns at that reference position. |
| Ref Base | The reference sequence base in this position. For multi-base deletions, the reference sequence of the string is shown, beginning with the base at the Ref Pos coordinate. If there is no reference sequence present, the Ref Base column displays the most frequently occurring non-ambiguous base at this position. If no such base exists, the consensus base at this position is shown. |
| Ref File | File name of the reference sequence used in the assembly. |
| Ref ID | Reference sequence or chromosome. |
| Ref Length | Length of the reference sequence used in the assembly. |
| Ref Name | The NCBI accession number for the short-read reference sequence. |
| Ref Pos | Reference position that does not include gaps. Coordinates matching entries in the VCF Variant table are shown. For deletions, Ref Pos is the genomic coordinate of the first deleted base. For insertions, Ref Pos is the genomic coordinate of the base preceding the insertion. |
| Region Capture | Indicates whether the variant occurs within a region specified in the .bed or manifest file used. Values are Yes and No. |
| Residue Count (A Cnt, etc.) | The number of bases of this type called in the aligned column. A dash (-) represents the reference base. |
| Sample | The sample name. |
| Splice | Variant is in or near an exon splice site. Splice site variations are changes to the 5’ (“donor”) or 3’ (“acceptor”) consensus splice site sequences. The DNA sequence for the donor is 5’-AGGTRAGT-3’ and for the acceptor is 5’-YYYYYYYYCAGGT-3’. Note that the AG dinucleotide on the 5’ end of the donor and the GT dinucleotide on the 3’ end of the acceptor are within the exon. Therefore, changes at these positions can also cause changes in the amino acid sequence of the resulting proteins. Changes in the intron portion of the splice site are marked as “Splice” in the column while those in the exon portion of the site are labeled “Splice in CDS.” Only the position where the change occurs is considered, not the identity of the base. |
| SNP | A manual evaluation score for the SNP, with a question mark being the default. See Change the status of a selected variant in the table below for a legend and instructions. |
| SNP % | The percentage of the sequence at this position in the assembly which varied from the reference. |
| Template Match ID | (DNASTAR internal use only). |
| Trace% | (DNASTAR internal use only) |
| Transcript ID | Transcript ID number from ENSEMBL. |
| Type | Specifies the variation type as SNP, Del (deletion) or Ins (insertion). For .assembly files and certain SQD files (e.g., from de novo or special templated workflows), pink typeface may be used to indicate non-synonymous variants. |
| User ID | Positions corresponding to a custom VCF Variant Table are labeled with the ID from that set. |
Need more help with this?
Contact DNASTAR



