From the Structural Variation view, click the Filter tool (
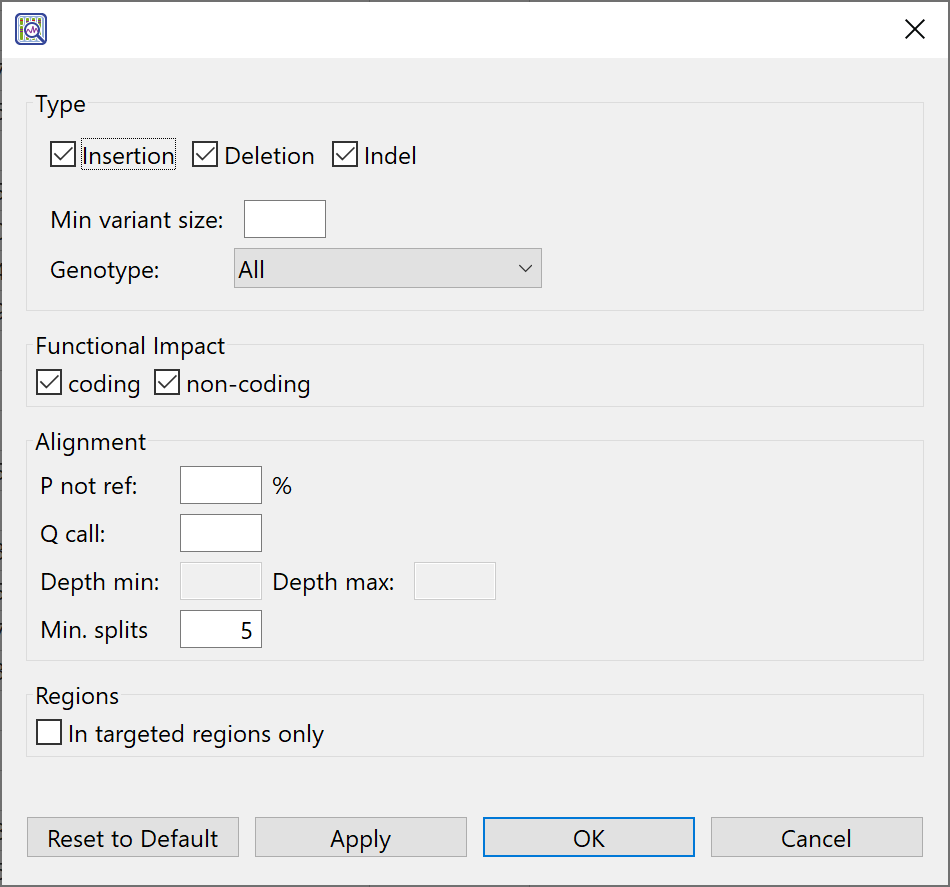
All settings in this dialog are optional, and it is fine to leave boxes blank if you do not wish to specify a threshold for that value. If desired, you can:
- Check or uncheck the boxes next to Ins, Del and Indel to show/hide insertions, deletions, or both types (indels).
- Specify a minimum threshold for variant length by entering a number in the Min. variant size box.
- Just as with SNPs, genotypes for structural variations may be heterozygous or homozygous. Use the Genotype drop-down menu to make the desired selection.
- Heterozygous Reference – To include SVs where one allele that differs from the reference and one that matches the reference.
- Homozygous Reference – To include SVs where both alleles match the reference.
- Reference – To include “possible” SVs; locations where there is evidence to suggest an SV even though Bayesian statistics show that both alleles most likely match the reference at that location.
- All – To include all of the above in the table.
- Heterozygous Reference – To include SVs where one allele that differs from the reference and one that matches the reference.
- Check or uncheck the boxes it you want to show/hide variations occurring in coding and non-coding regions.
- Enter a percent threshold for P not ref, the probability that the called base at this position is not the reference base.
- Enter a threshold value for Q call, the Phred-like quality score of the called genotype. Q call is a measure of the probability that the called genotype is correct.
- To the right of Min splits, enter the minimum number of “split reads” needed to determine the breakpoints of a deletion.
- Check the In targeted regions only if you specified targeted regions during assembly with SeqMan NGen and only want to see variations in those regions.
Use the buttons at the bottom of the screen to apply changes to the Structural Variations view and/or close the wizard.
- Use Apply to apply the current wizard selections to the view while keeping the wizard open.
- Use OK to apply the current wizard selections to the view and to close the wizard.
- Use Cancel to close the wizard without applying changes to the view.
Saving filters and applying saved filters to future views:
| Task | How To |
|---|---|
| Save the current filter settings to apply (manually) to future Structural Variations views | At the top right of the Structural Variations view, click on the Filter tool ( ) and choose Save Filter. ) and choose Save Filter. |
| Save the current filter settings to apply as the default for all future Structural Variations views | At the top right of the Structural Variations view, click on the Filter tool ( ) and choose Save Filter as Default. ) and choose Save Filter as Default. |
| Apply filter settings that were saved previously | At the top right of the Structural Variations view, click on the Filter tool ( ) and choose Apply Filter. ) and choose Apply Filter. |
| Restore the DNASTAR default filter settings | At the top right of the Structural Variations view, click on the Filter tool ( ) and choose Restore DNASTAR Default Filter. ) and choose Restore DNASTAR Default Filter. |
Need more help with this?
Contact DNASTAR


