To navigate to a specific position:
Do any of the following:
- Open the Places panel and double-click on a previously-stored location.
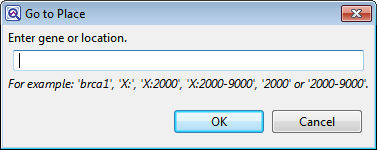
- Use Edit > Go to Place or press Ctrl/Cmd+J. In some cases, the right-click option Go to Place may also be available. In the Go to Place dialog, type the desired feature name, numerical position, or numerical range. Then click OK.
To navigate to a location or feature using the Search bar:
The search tools are available from the Overview and Analysis view. By default, the search tools are hidden.
- Display the tools by clicking the Find tool (
) at the top right of either view or use Edit > Find. This causes the search tools to be displayed on the top left of that view.

- Use the Find drop-down menu to choose the item to search for. Available search target options depend on the project type (.sqd vs. .assembly), assembly type (de novo vs. templated), and the view (Analysis view vs. Overview). The table below shows possible options in alphabetical order.
| Search Target | Description |
|---|---|
| Coverage | Search for the edge of a region that is below the minimum threshold coverage. |
| Go to alignment position | Go to a specific numerical position on the aligned sequence rows. |
| Go to reference position | Go to a specific numerical position on the reference sequence. |
| Reference feature | Searches for and selects the next or previous feature on the reference sequence. To see these features in the view, be sure to check the Reference > Features box(es) in the Tracks panel. |
| Reference feature by name | Search for and select a feature by entering its name and by optionally limiting the search to particular types, such as gene, CDS, etc. |
| Region | Search for genomic regions, if a BED file is included in the session; or phase blocks, if the session includes phased data. |
| Variant | Locates bases that have been identified by SeqMan NGen’s SNP-calling detector as variants. Variant is available for reference-guided assembly projects only. |
- Different search options may cause additional drop-down menus or text boxes to appear, allowing you to type in a text string of query bases, choose a feature category, etc. In some cases, a box is provided to Allow ambiguity. Check the box if you want to include IUPAC ambiguity codes in your search.
If you type a query that is “too wide” for the search box, you can hover over the search box to view the entire query. Note that long query sequences may take considerable time to locate.
- Use the green right/left arrows (
) or the Edit > Find Next and Edit > Find Previous commands to jump to the next/previous find. After reaching either end of an alignment, GenVision Pro wraps around and continues searching from the other end. Each time a feature is located, it is selected in all views until you press one of the green arrows again.
Need more help with this?
Contact DNASTAR



 ) at the top right of either view or use Edit > Find. This causes the search tools to be displayed on the top left of that view.
) at the top right of either view or use Edit > Find. This causes the search tools to be displayed on the top left of that view. ) or the Edit > Find Next and Edit > Find Previous commands to jump to the next/previous find. After reaching either end of an alignment, GenVision Pro wraps around and continues searching from the other end. Each time a feature is located, it is selected in all views until you press one of the green arrows again.
) or the Edit > Find Next and Edit > Find Previous commands to jump to the next/previous find. After reaching either end of an alignment, GenVision Pro wraps around and continues searching from the other end. Each time a feature is located, it is selected in all views until you press one of the green arrows again.