The Variants view shows a table with a row for each variant position and can only be generated for reference-guided assemblies in .assembly format. Depending on which command or tool is used to access the view, the table may contain all variants, filtered variants, or a set of variants that was saved previously. In addition, the Variants view comes in two versions: single-sample and multi-sample.
For a general overview with a table that shows how to recognize different versions of the Variants view, see Explore Variants and Structural Variations.
To learn how to generate a view, perform filtering, create variants sets, use the header tools, and much more, see:
- Compare variants across samples / Filter a multi-sample Variants view
- Analyze variants in a single sample / Filter a single-sample Variants view
To learn how to work with variant sets, see Create and analyze variant sets.
The remainder of this present topic covers only information not provided in the topics just described.
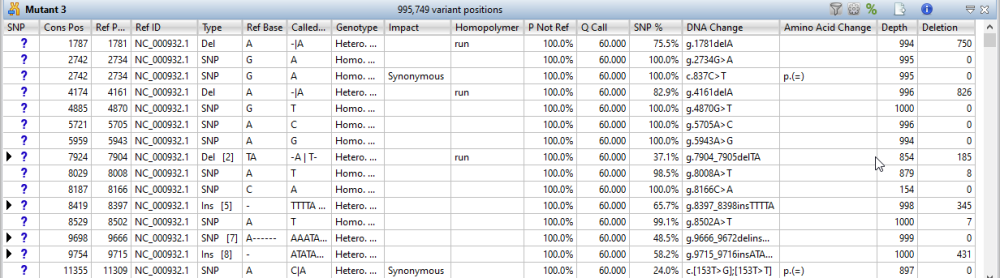
Each row in the table summarizes information for all of the variant bases in an aligned column. If you are viewing a SeqMan NGen assembly, only variants meeting the SNP filter stringency (High, Medium or Low) that you specified in the SeqMan NGen wizard are displayed. For definitions of each data column, as well as instructions for showing, hiding, renaming or rearranging data columns, see Choosing and understanding Variants view columns.
The table below describes some tasks related to this view that are shared in both versions of the view and are not included in the linked topics above.
| Task | How To |
|---|---|
| Open the Analysis view at the variant position | Click on any row in the Variants view to snap to the location in the Analysis view. This is especially useful in situations such as an amplicon assembled against a much longer reference. |
| Select variants in a particular region of the sequence | Drag your mouse across a region in the Analysis view to select the region’s variants in the Variants view.  |
| Change the status of selected variants | The leftmost column of the table (represented by a small triangle in the header) denotes the status of a variant. Initially, all variants are marked as putative.
|
| Hide/show multiple references bases | If there were multiple reference bases at the location of a variant, the leftmost column will feature a small triangle in that row. Click on the triangle to show the multiple reference bases.  In .assembly projects, variants in adjacent columns are coalesced into a single insertion or deletion if they are of the same type, and if at least 80% of the reads with the called variant in one column have a variant in the adjacent column. Each coalesced multiple-base insertion or deletion can be opened to reveal individual variants by clicking the corresponding triangle to the left of the SNP column. After clicking a triangle, information on each position of the insertion/deletion is displayed in a separate row.  |
Need more help with this?
Contact DNASTAR



 – Putative (the initial status for all variants in the table)
– Putative (the initial status for all variants in the table)  – Confirmed
– Confirmed  – Rejected
– Rejected