In NovaFold, protein fragments from existing PDB structures are used as templates for building a prediction model. The Templates section of the NovaFold Report lets you compare the sequence similarity of a query sequence to the top ten template sequences in MegAlign Pro.
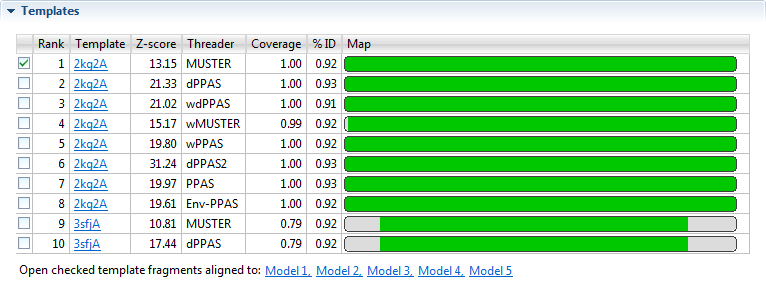
- Rank - Displays a list of the top ten threading templates, with “1” representing the best template. Use the checkboxes at the left of the Rank column to specify which template(s) to align against a selected model (see bottom of this row).
- Template - The name of the template with a link to the entry on the PDB website.
- Z-score - A normalized score describing the similarity between the query and the template. Higher is better. A score ≥ 1.0 is considered to be significant.
- Threader - Threading algorithm that selected the template. NovaFold uses eight different algorithms.
- %Coverage - Fraction that the template overlaps the query sequence (0,1].
- %ID - The percentage of the query sequence that is an exact match to the template sequence. Templates with low sequence similarity may still be structurally similar to the query sequence, but selecting these relevant templates is challenging.
- Map - A minimap showing the alignment of the query sequence to the template.
- Green - ID ≥ 70
- Amber - 30% ≤ ID <70
- Red - ID <30
- Gray - gaps
- Green - ID ≥ 70
Check one or more templates, and then click any of the links entitled “Open checked template fragments aligned to Model ‘n’” to open a new Protean 3D project showing each of the checked templates aligned with the selected model. In the Protean 3D document containing the requested alignment, each protein fragment will appear as an entry in the Features panel, and as a region feature in the Analysis view.
The next section of the NovaFold Report is the Model Overview section.
Need more help with this?
Contact DNASTAR


