If you checked Predict ligand binding sites and protein function in the NovaFold Options screen, the Predicted Binding Sites section of the NovaFold Report is available for Model 1, and for any model with a predicted TM-score ≥ 0.5. This section lists predicted ligand binding sites for a NovaFold prediction in a model structure.
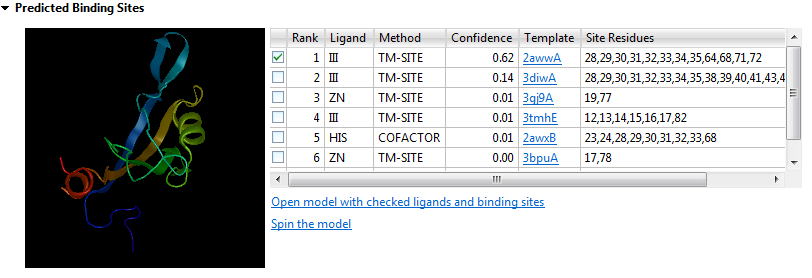
The thumbnail image is a mini Structure view that shows a rendering of the predicted structure and the ligands checked in the Rank column of the table.
- Rank - Orders the ligands that the query sequence is likely to bind to from one to ten, with one being the most likely. Use the checkboxes at the left of the Rank column to specify which ligands to include as features when opening the model structure (see bottom of this row).
- Ligand - Abbreviation for the ligand name.
- Method - The proprietary algorithm used in the calculation: TM-SITE, COFACTOR or S-SITE.
- Confidence - Confidence score [0,1], with higher numbers denoting increased confidence.
- Template - The name of the template with a link to the entry on the PDB website.
- Site Residues - The numbers of the residues involved in the predicted binding site.
There are two links at the right:
- Click Open model with checked ligands and binding sites to open the model structure with each of the checked ligands as Features in the Features panel. This allows you to view similarities between a predicted binding site and known binding sites.
- Click Spin the model to spin the model left. When the model is spinning, you can stop it at any point by clicking Stop spinning the model.
In certain cases, the next section of the NovaFold Report is Predicted Enzyme Function.
Need more help with this?
Contact DNASTAR


