To design primer pairs:
Open the Sequence, Linear, Circular or Minimap view and use the mouse to select the region you wish to amplify. If none of the sequence is selected, the entire sequence will be amplified. Choose Priming > Create Primer Pairs.
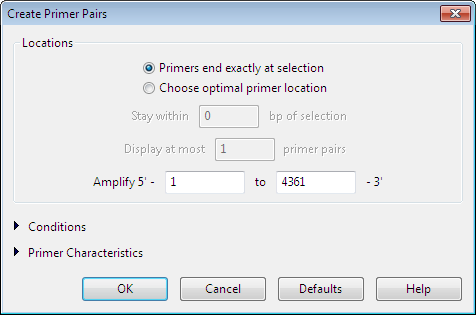
In the Create Primer Pairs dialog, define the required conditions for identifying primer candidates.
Locations settings:
The Locations section of the Create Primer Pairs dialog is expanded by default and allows you to specify parameters that define where SeqBuilder Pro will search for primers on your sequence.
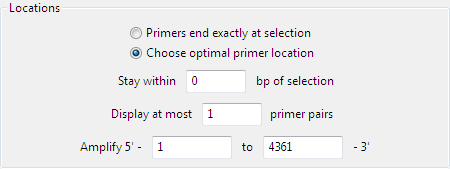
Specify allowable primer locations by selecting one of the two radio buttons:
- Primers end exactly at selection searches for primers that lie exactly within the selected region.
- Choose optimal primer location searches for a specified maximum number of primers that begin and end within the distance specified outside of the selected region. When this option is selected, two additional options are enabled. The defaults are to Stay within 0 bp of the selection and Display at most 1 primer pair. If desired, you may type different numbers in these boxes.
The region selected for amplification can be changed by editing the coordinates in the Amplify 5’ to 3’ fields. If you wish to create a product that wraps the origin, enter a 3’ coordinate that is less than the 5’
coordinate. Alternately, you can select a feature that wraps the origin prior to opening the dialog. To create product on the reverse strand, select the region you wish to amplify as a feature on the reverse strand before opening the dialog.
Conditions settings:
The Conditions section of the Create Primer Pairs dialog allows you to specify the initial salt concentration, which affects the calculation of predicted melting temperature. This section is minimized by default and can be expanded by clicking on the small arrow to the left of Conditions.

By default, the Salt concentration is 50.0 mM. This value may be adjusted to correspond to actual experimental conditions.
Primer Characteristics settings:
The Primer Characteristics section of the Create Primer Pairs dialog enables you to limit your search for primers based on primer length, target melting temperature, and primer interactions at the 3’ end. You also have the option to avoid known repetitive sequences that are likely to cause mispriming. This section is minimized by default and can be expanded by clicking on the small arrow to the left of Primer Characteristics.
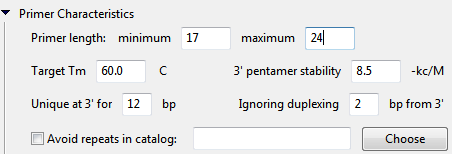
- Primer length minimum defines the minimum acceptable primer length. Primers must be at least this length (in bp) in order to be considered for selection.
- Primer length maximum defines the maximum acceptable primer length. Primers must not exceed this length (in bp) in order to be considered for selection.
- Target Tm defines the target primer melting temperature. SeqBuilder Pro will attempt to locate primers as close to this value as possible.
- 3’ pentamer stability defines the threshold for pentamer stability at the 3’ end of the primer. Pentamer dG values at the 3’ end exceeding this value will not be considered for selection. See The Pentamer Profile for additional information.
- Unique at 3’ for n bp defines the number of base pairs at the 3’ end of the primer that must be unique in order to be considered for selection. Uniqueness of at least 12 or more bases reduces the likelihood of mispriming at another site.
- Ignore duplexing n bp from 3’ defines the number of base pairs from the 3’ end of the primer where duplexing can be ignored.
- Check the box next to Avoid repeats in catalog if you have known repetitive sequences that you wish for SeqBuilder Pro to ignore when selecting primer locations. Click Choose to locate and select your file containing the repetitive sequences. The file must be a tab-delimited .txt file created manually through a text editor. If a .txt file is used, each line should include, from left to right, the sequence fragment, then a tab, then the name or notation.
______________________________
After making changes, you can return to the original settings, if desired, by clicking the Defaults button. Once the settings are as you wish, click OK to begin the search.
After attempting to create primer pairs automatically, there are two possible outcomes:
- If primer pairs were not located, a dialog appears saying that “No primer pairs were found.” In most cases, this issue can be resolved by changing the settings in the Primer Characteristics dialog. This time, allow SeqBuilder Pro to choose the primer locations by using or relaxing the Choose optimal primer location parameter. Alternatively, you can create primers using Priming > Create Primer from Selection and then manually pair them together.
- If primer pairs were located, SeqBuilder Pro creates and displays a PCR_product feature, two PCR_primer features, and a PCR_frame feature for each located primer pair. Just like any other feature, these are visible in the Primer Design view and the graphical views.
Need more help with this?
Contact DNASTAR


