To modify and rename a genetic code:
- Open a sequence or SeqBuilder Pro project.
- Select Sequence > Edit Genetic Code. The two-tabbed Genetic Code Editor appears.


| Task | How to… |
|---|---|
| To assign a name for the code | Type a name into the Description field. Warning: If you do not make changes to this field, the original genetic code will be overwritten with any changes made below. |
| To specify whether the code is for RNA or DNA | Select RNA or DNA from the drop-down menu in the upper right of the dialog. |
| To modify codon assignments, including stop codons | In the Genetic Code tab, use the mouse to drag a codon to the appropriate amino acid row. The color of the codon will change from red to black to show it has been moved. SeqBuilder Pro only considers red codons when it calculates the aggregate codon for the set. |
| To include the newly moved codon when calculating the aggregate codon | In the Genetic Code tab, Alt+click (Win) or Option+click (Mac) on the codon to change it back to red. The sum of all codons in the row is recalculated to give the revised reverse translation codon. The same recalculation occurs instantly when a codon is removed from a row. Reverse translation codons, shown in the third column from the left, are the codons used in reverse translating a protein sequence. |
| To change the codons that will be used for back translation without changing the forward translation | In the Genetic Code tab, Alt+click (Win) or Option+click (Mac) on codons to exclude. For example, in the Standard Genetic Code, there are two codons for Cysteine (UGU and UGC) and the default reverse translation codon is UGY (Y means pyrimidine, i.e. C or U). To reverse translate Cysteine residues as UGC, Alt+click (Win) or Option+click (Mac) on the UGU codon to change its color to black so it is not considered for reverse translation. The reverse translation codon for Cysteine will now read UGC instead of UGY. |
| To edit start codons | In the Start Codons tab, select the desired start codons visually by finding the row containing the first base (on the left), following that over to the column belonging to the second base (in the center) and selecting the row in the table that aligns with the third base (on the right). Click an amino acid to turn it green, and cause > to appear beside it. The codon representing the amino acid now appears at the top of the screen to the right of the word Starts. |
| To remove a start codon from the list at the top of the page | In the Start Codons tab, click on the corresponding amino acid in the body of the table. |
- Click OK. If you have changed the description, SeqBuilder Pro requests confirmation.
| Task | How to… |
|---|---|
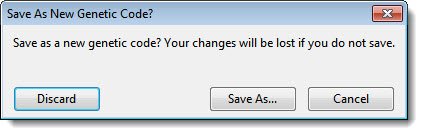
| To refuse the changes and return to using the original configuration of the genetic code | Select Discard. |
| To save the genetic code under a new name | Select Save As. In the Save dialog, type a name for the modified code. Do not change the save location. Press Save. |
| To return to the Genetic Code Editor and continue modifying the genetic code | Select Cancel. |
Need more help with this?
Contact DNASTAR


