The Peak Detection tab is used to set parameters for MACS peak detection. To access the tab from the Analysis Options screen, click the Advanced Analysis Options button then click on the Peak Detection tab. The options available in this tab may vary depending on the workflow.
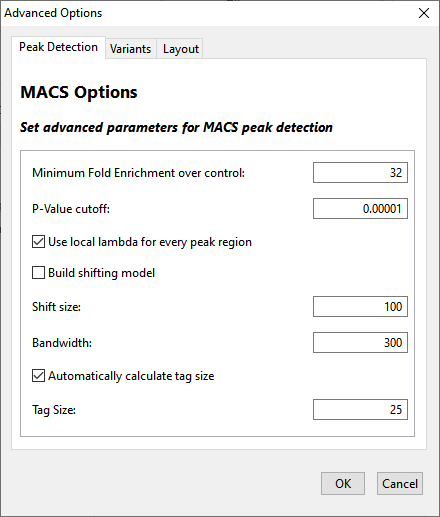
| Parameter | Description |
|---|---|
| Minimum Fold Enrichment over control | This parameter controls how enriched a peak must be, compared to the background read distribution, in order to be considered in building the peak model. If a previous assembly attempt returned the error message “too few paired peaks to build a model,” we recommend using a lower number in this box. |
| P-Value cutoff | Local read distribution is compared to the threshold value entered here in order to calculate whether a peak should be counted as “real.” SeqMan NGen calculates the likelihood that a detected peak is actually a peak based on the local read distribution and only returns peaks with values below the P-Value cutoff. The default P-value cutoff is 0.0001. This doesn’t mean that values greater than 10-5 are filtered out, but rather that they are not included in the count of “real” peaks. |
| Use local lambda for every peak region | Lambda is a parameter used to define a Poisson distribution which MACS uses to determine the expected number of reads in a given region. When this option is checked, the Poisson distribution is calculated for the peak region and for three regions surrounding the peak. MACS can use this information to determine the expected number of reads in a given region. If the option is unchecked, then the local distributions are not calculated. Instead the expected distribution is based on the total number of reads and the effective size of the genome. |
| Build shifting model | Check this option to have MACS build a model based on the data to determine the width of and the distance between the "paired peaks.” Alternatively, leave this option unchecked to set Shift Size and Bandwidth values manually. The Shift Size is the distance each of the paired peaks will be shifted to try to center them over the actual binding site. The Bandwidth value defines the expected width of peaks. SeqMan NGen will search for peaks using a window twice as long as the bandwidth. Note: The creators of the MACS algorithm advise disabling model building when dealing with broad peaks (i.e. binding sites); for example, when studying histone binding. |
| Shift size | This parameter is described in Zhang et al., 2008. |
| Bandwidth | This parameter is described in Zhang et al., 2008. |
| Automatically calculate tag size / Tag Size | MACS treats all reads as though they have equal length. To explicitly specify that length, check Automatically calculate tag size and enter a length in the Tag Size text box. |
Need more help with this?
Contact DNASTAR


