In most cases, de novo sequence assembly will yield multiple contigs rather than a “perfect” single contig. DNASTAR offers several methods for filling in (AKA “closing”) the gaps between contigs:
- SeqMan NGen supports a Hybrid reference-guided/de novo genome assembly workflow. For information on this method, see the SeqMan NGen help.
- SeqMan Ultra offers three ways to close gaps, as shown in the following video. One of these methods is described step-by-step in the section below the video.
To close a gap between two contigs in SeqMan Ultra:
The following steps describe the most popular method for closing gaps using SeqMan Ultra. For other methods, see the video above.
Before you begin, note that this method works only for .sqd files (not .assembly files) that contain mate-pair data.
- Open an .sqd assembly project in SeqMan Ultra.
- Order and scaffold contigs using either of these methods:
- Automatically: Choose Contig > Order Contigs. SeqMan Ultra will use mate-pair data to create one or more scaffolds consisting of multiple contigs already pre-numbered in increasing order.
- Manually: Use mate-pair data in the Strategy view to discover the relationship between contigs. When you find two or more contigs that you believe go together, select them in the Explorer panel and choose Contig > New Scaffold. Manually type their relative position numbers into the Position column. Choose any novel number (i.e. a number not already used in the Explorer panel) for the first contig, and larger numbers for each successive contig.
- Automatically: Choose Contig > Order Contigs. SeqMan Ultra will use mate-pair data to create one or more scaffolds consisting of multiple contigs already pre-numbered in increasing order.
- Select the scaffold in the Explorer panel and open the Strategy view.

- Find the end/beginning of two contigs you wish to join. You can skip from one contig to the next by setting the search type to Contig and using the blue arrow keys.

- When you find a gap you would like to close, use your mouse to select a range of sequence near the end of either contig.
- Choose Search > Search to initiate a BLAST search of that contig end. Choose the desired settings and press Run Now or Run.
- Once the job finishes, click the link in the Status column of the Jobs panel to open the search results.
- Click on the best match, then right-click and choose Download Sequences.

- In the Explorer panel, select the two contigs on either side of the gap and choose Contig > Add Sequences to Close Gap.
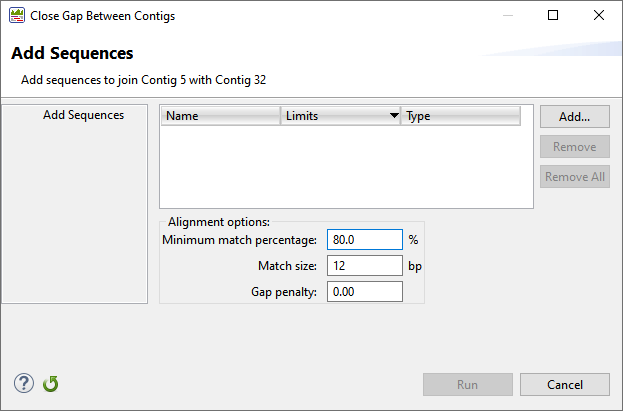
- Use the Add button to upload the saved match, adjust the alignment options as desired, and click Run.
A report shows details of the gap closing attempt, including whether or not it appeared to be successful.
- Once you complete a successful gap closure, open the Strategy view to view the closed gap.
- If there are additional gaps, follow the same steps to close each of them.
- Once you have closed as many gaps as possible, export the consensus to use in setting up a new templated assembly in SeqMan NGen.
Need more help with this?
Contact DNASTAR


