After the Jobs panel shows the Status of a BLAST search or Entrez search is Done, pressing the “Done” hyperlink opens the search results simultaneously in the Text, Table, and Pairwise views. All three views are contained in a large pop-up window that is separate from the main SeqMan Ultra window.
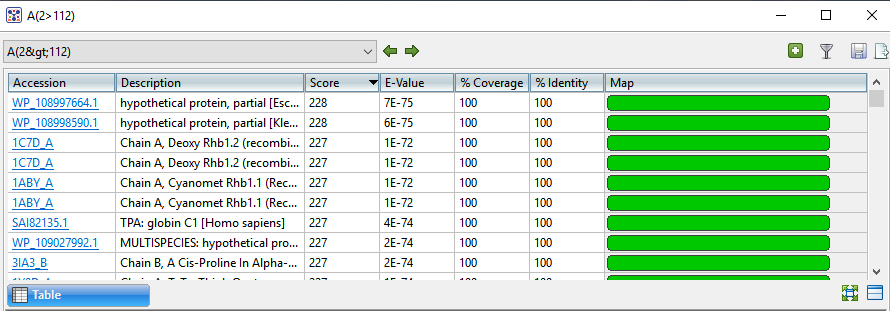
If no significant matches were identified, the Table view will be empty. An example of a populated Table view is shown below:
Columns in the view’s table vary according to the search type. Some of the possible columns are described below:
| Column | Description |
|---|---|
| Accession | The NCBI “Accession” number for the match. Press the link to open the corresponding page on the NCBI website. |
| Description | The NCBI “Definition” field text. |
| Length | The length of the sequence in the match. |
| Organism | The name of the organism associated with this match. |
| Score | The similarity score for the match. (See the NCBI definition for raw score). In general, a higher score denotes a better match. |
| E-Value | The chance that the match is expected to occur by chance. (See the NCBI definition for E value). In general, a lower score denotes a better match. |
| %Coverage | The percentage of the query sequence that aligns to a match in the BLAST database. Example: Query has 596 bases, of which 565 align to a match in the NCBI database. %Coverage = 565/596 = 95%. |
| %Identity | The percentage of bases that match when the query and result sequence are aligned. (NCBI definition). Example: Query has 565 bases that align to the match and 3 bases that don’t match. %Identity = 562/565 = 99.5%. |
| Map | Image showing areas of agreement between the query and the match. |
Tasks pertaining to the Table view:
| Task | How To… |
|---|---|
| If a BLAST or Entrez search was performed | |
| Launch the NCBI page for matching database entry | Click the hyperlink in the table’s Accession column. |
| Copy all information from a search result row | Right-click on the row and choose Copy. |
| Save the sequence match in .gbk format | Right-click on the row and choose Download sequences. In the ensuing Save As dialog, choose a location in which to save the sequence. By default, the sequence is named using its Accession number. |
| Add sequences or matches | Use the Add sequences or matches tool ( ). Sequences are added back into the host application, not the job’s document. ). Sequences are added back into the host application, not the job’s document. |
| Filter results from a sequence or text search | Use the Filter results tool ( ). Learn more about sequence search or text search filtering. ). Learn more about sequence search or text search filtering. |
| Export data from the view | Use the Export data tool (  ), described in the topic Export data from the Text, Table or Pairwise views. ), described in the topic Export data from the Text, Table or Pairwise views. |
| Only if a BLAST search was performed | |
| To save results | Click the Save as tool ( ). A save dialog opens and allows you to save the search results as a .search format file. This option is not available for Entrez searches. ). A save dialog opens and allows you to save the search results as a .search format file. This option is not available for Entrez searches. |
| To display a pairwise alignment using a particular search result | If both the Table and Pairwise views are open, click on the desired row in the Table view to see the corresponding pairwise alignment in the Pairwise view. As an alternative, open the Pairwise view manually and choose the desired target from the with drop-down menu. |
Need more help with this?
Contact DNASTAR