GenVision
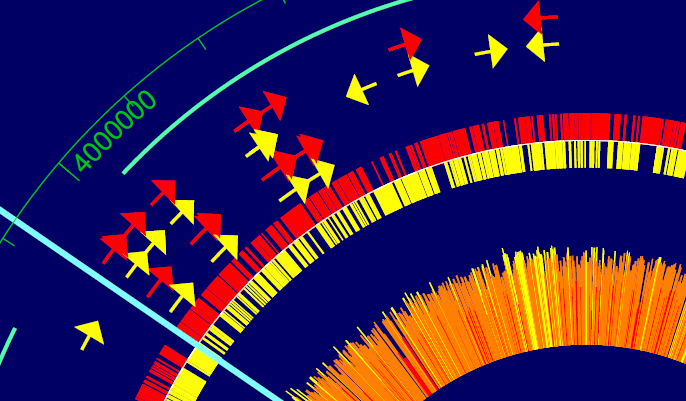
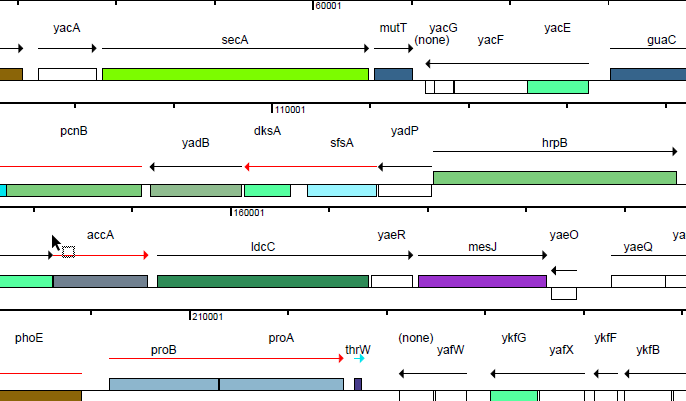
When it comes to illustrating your genomic data for a publication or presentation, a high-resolution map is worth a thousand words. GenVision is a genomic visualization application that supports the generation of publication-quality graphics for linear or circular genome mapping. GenVision can be customized to accentuate specific areas of interest in your annotated genome, enabling the graphic comparison of gene functionality, illustration of gene expression levels, or visualization of the coverage in an assembled contig.

UNIVERSITIES TRUST DNASTAR
BIOTECH AND PHARMA COMPANIES RELY ON DNASTAR
JOURNAL CITATIONS OF DNASTAR
DNASTAR USERS WORLDWIDE
Resources
Please see our resources below for more information on how to create an annotated genome in GenVision.
Creating a Genomic Map
Using Data Files to Build a Genome Map
Working With Panels in GenVision
Integrating Data into a Single Illustration
GenVision Help
GenVision Tutorials
FAQs
Which Lasergene package includes GenVision?
For Windows users, GenVision is included in Lasergene Molecular Biology as well as in the complete DNASTAR Lasergene package which includes all of the applications from Lasergene Molecular Biology, Lasergene Protein, and Lasergene Genomics…
For Windows users, GenVision is included in Lasergene Molecular Biology as well as in the complete DNASTAR Lasergene package which includes all of the applications from Lasergene Molecular Biology, Lasergene Protein, and Lasergene Genomics.
The full GenVision application is available on Windows only, but macOS users can view, convert and print GenVision projects using the GenVision Utility, which is installed automatically as part of every Lasergene license.
How do I create an annotated genome map in GenVision?
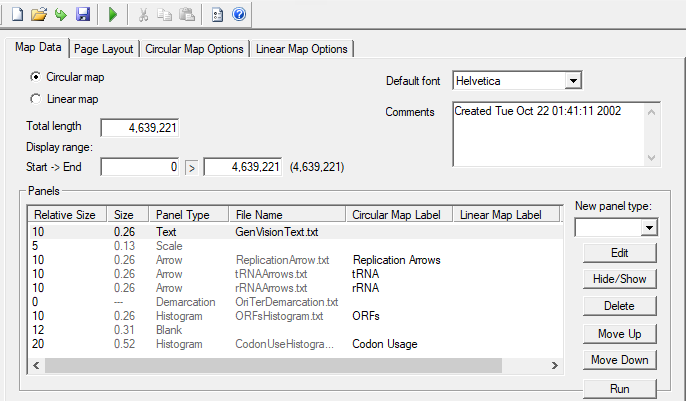
GenVision illustrations are built from one or more panels, each drawn using information from an associated data file. Data files contain the annotation data you wish to display in a tab-delimited format, and may be created manually, in a program such as Microsoft Excel, or automatically by exporting from a Lasergene application…
GenVision illustrations are built from one or more panels, each drawn using information from an associated data file. Data files contain the annotation data you wish to display in a tab-delimited format, and may be created manually, in a program such as Microsoft Excel, or automatically by exporting from a Lasergene application. There are several different types of panels that you may use to visualize your annotation data, including arrows, histograms, graphs, and text.
What is the difference between GenVision and the GenVision Utility?
The full GenVision application is available only on Windows, but both Windows and macOS users can view, convert and print GenVision projects using the GenVision Utility, which is included with every Lasergene installation…
The full GenVision application is available only on Windows, but both Windows and macOS users can view, convert and print GenVision projects using the GenVision Utility, which is included with every Lasergene installation.
The GenVision Utility can be used to:
- Convert a saved GenVision project to Postscript format.
- Convert a GenVision project to PDF format and view the image.
- Print a saved GenVision project image.
Compare DNASTAR Lasergene Packages
| MOST POPULARDNASTAR Lasergene | ||||
| Lasergene Molecular Biology | Lasergene Genomics | Lasergene Protein | ||
|---|---|---|---|---|
| Included Applications | ||||
| SeqBuilder Pro | ||||
| SeqMan Ultra | ||||
| MegAlign Pro | ||||
| GeneQuest | ||||
| GenVision | ||||
| SeqNinja | ||||
| SeqMan NGen | ||||
| ArrayStar | ||||
| GenVision Pro | ||||
| Protean 3D (+1 prediction per Nova Application) |
||||
| DNASTAR Navigator | ||||
| Supported Workflows | ||||
| Integrates with | ||||
| See Pricing | See Pricing | See Pricing | See Pricing | |