MegAlign Pro
MegAlign Pro offers a complete sequence alignment software package, including everything you need for each stage of an alignment – not only the algorithms needed for aligning both gene-level and genome-scale sequence data, but also the capability to dig deep in the post-alignment stage.
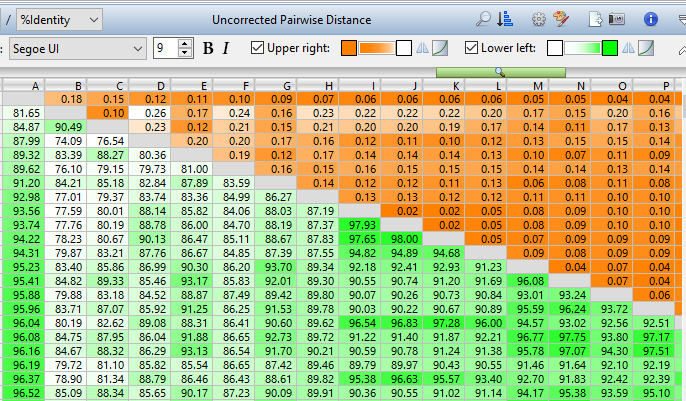
MegAlign Pro performs multiple and pairwise sequence alignments quickly and easily, then guides you through the post-alignment process, allowing you to generate phylogenetic trees, evaluate variants, separate interesting regions for new subalignments, edit and trim individual sequences or the entire alignment, and customize the appearance of your alignment before generating high-quality images, suitable for publication.

UNIVERSITIES TRUST DNASTAR
BIOTECH AND PHARM COMPANIES RELY ON DNASTAR
JOURNAL CITATIONS OF DNASTAR
DNASTAR USERS WORLDWIDE
MegAlign Pro Workflows
Resources
Please see our resources below for more information on MegAlign Pro sequence alignment software.
Gene Homology at Scale
Simplifying Multiple Sequence Alignment and Analysis with MegAlign Pro
Comprehensive Variant Analysis: from Raw Sequence Data to Aligned Genomes
Master Viral Genome Analysis Webinar
Mastering Phylogenetic Tree Creation & Optimization with MegAlign Pro
How to Create the Best Phylogenetic Tree for Your Data Using MegAlign Pro
Two ways to find the best MegAlign Pro multiple sequence alignment method for your data
Using the MAFFT alignment algorithm for high-capacity viral genome alignment
FAQs
Which Lasergene package includes MegAlign Pro?
MegAlign Pro is included in Lasergene Molecular Biology as well as in the complete DNASTAR Lasergene package which includes all of the applications from Lasergene Molecular Biology, Lasergene Protein, and Lasergene Genomics.
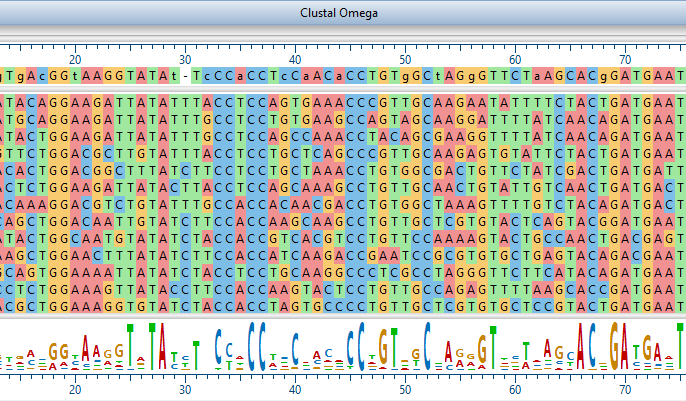
What multiple sequence alignment methods are available in MegAlign Pro?
MegAlign Pro sequence alignment software includes Clustal Omega, Clustal W, MAFFT, and MUSCLE algorithms for aligning both DNA and protein sequences.
Can I modify my multiple sequence alignment in MegAlign Pro?
Yes. MegAlign Pro supports multiple ways to modify an alignment. You can:…
Yes. MegAlign Pro supports multiple ways to modify an alignment. You can:
- Trim ends at the beginning or end of an alignment.
- Realign a range of aligned sequences using different parameters or a different alignment engine.
- Merge two existing completed alignments together.
- Add all/selected sequences from the “Unaligned Sequences” area into the current alignment, while retaining all of the gaps of the original, pre-merge alignment.
- Realign existing aligned sequences plus selected unaligned sequences
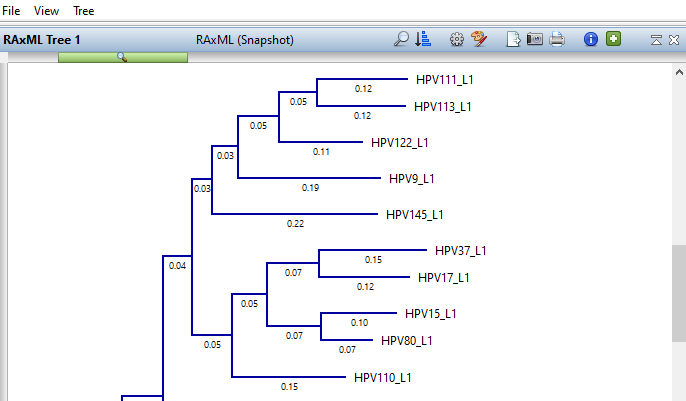
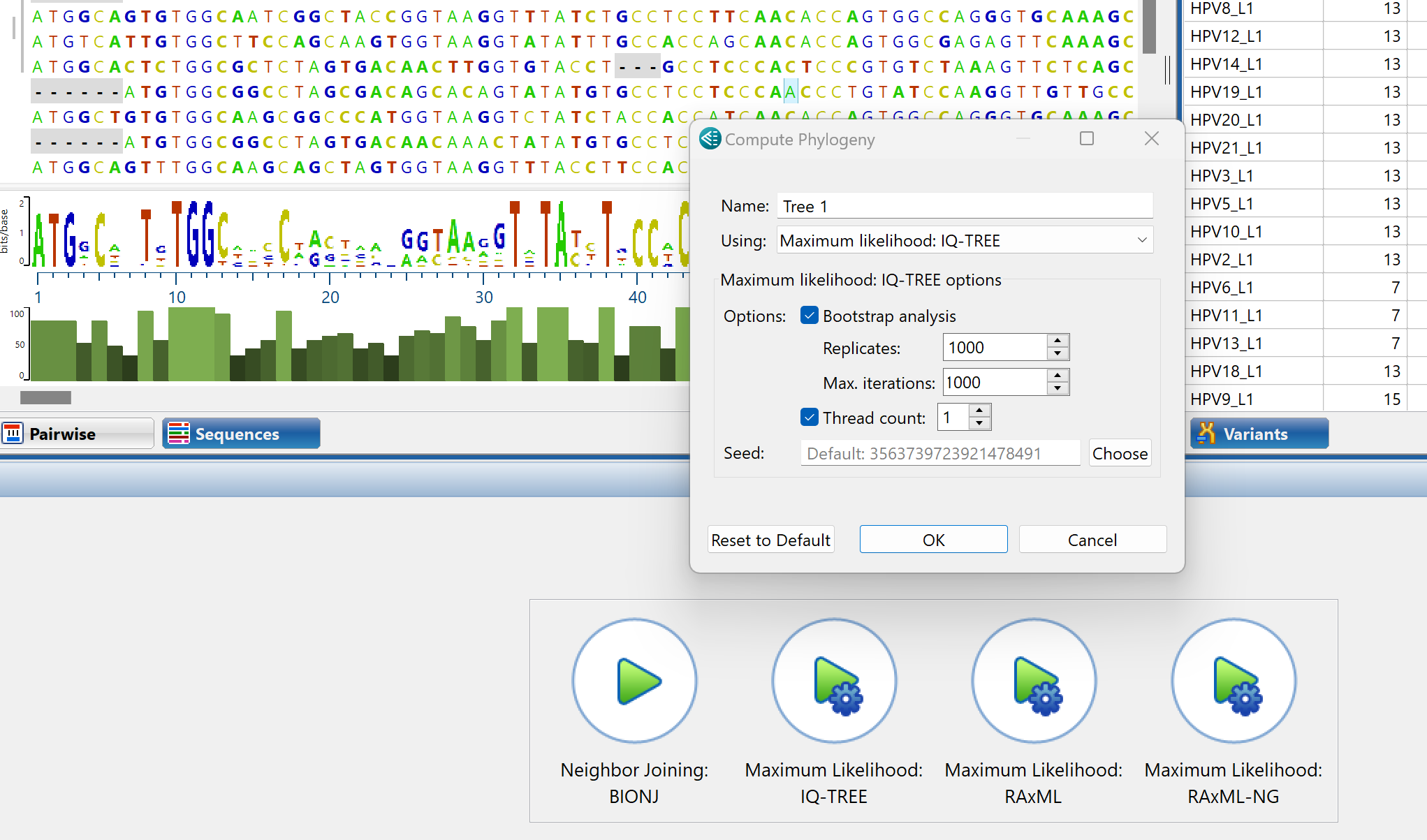
What methods does MegAlign Pro offer for computing phylogeny?
MegAlign Pro offers four methods for generating phylogenetic trees. RAxML, RAxML-NG, and IQ-Tree all use different versions of the Maximum Likelihood algorithm. The fourth tree generation method is Neighbor Joining, which uses the BIONJ algorithm.
Can I search NCBI databases online? Can I search local databases?
GenVision Pro, Protean 3D, SeqMan Ultra, SeqBuilder Pro, and MegAlign Pro all offer the ability to do online sequence similarity searches in NCBI BLAST and text similarity searches in NCBI Entrez.
In addition, all but SeqBuilder Pro offer the ability to create and search local databases. First, use the Local Database Manager to upload local files and/or import sequences from NCBI, with over 30 databases to choose from. The manager can also be used to merge, expand, rename, and/or delete local databases. Local searches can then be done without Internet access and are a great way to ensure data security. In addition, local searches are 5–25x faster than online searches.
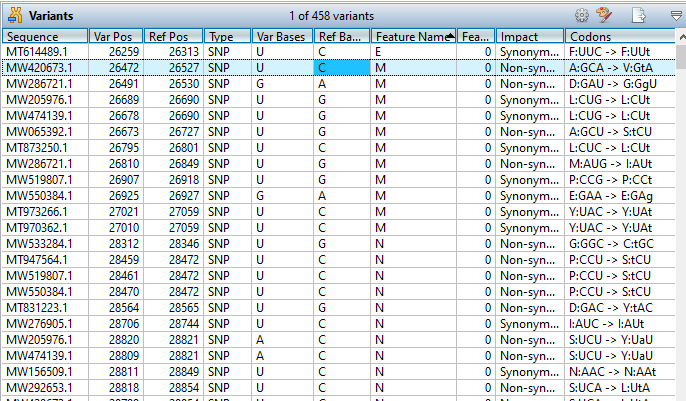
How can I evaluate the variants in my alignment?
MegAlign Pro provides extensive variant analysis capabilities for viral, mitochondrial, and chloroplast genomes. To analyze variants…
MegAlign Pro provides extensive variant analysis capabilities for viral, mitochondrial, and chloroplast genomes. To analyze variants, first specify a reference sequence, either before or after alignment. Then choose Variants > Compute Variants. Details about the variants in your alignment, including position, feature, impact, and translation changes, can be analyzed and evaluated in the Variants view.

What pairwise alignment methods does MegAlign Pro offer and how do I know which one to use?
MegAlign Pro features three pairwise sequence alignment tools:
- Local Pairwise Alignment is designed…
MegAlign Pro features three pairwise sequence alignment tools:
- Local Pairwise Alignment is designed specifically to find the highest scoring aligned segments of two sequences, even if the full extent of the two is not included in the final alignment.
- Global Pairwise Alignment doesn’t try to find the best scoring segment, but instead requires that the full extent of both sequences be included in their results. There is no requirement or guarantee that the best scoring pair of aligned segments from a local alignment will be aligned in a global alignment.
- Semi-Global Pairwise Alignment is a relatively new approach that is particularly suitable when the two sequences differ greatly in length. When that happens, the longer sequence will have overhangs on either end of the alignment. Since overhangs are represented with gaps, a global aligner will attempt to increase the match score and minimize accumulated gap penalties by aligning parts of the shorter sequence to overhanging sequence region(s). This effect can produce a number of unrealistic, usually small aligned segments spaced by gaps near the ends of the alignment. Semi-global alignment is designed to address this problem by not penalizing gaps in overhangs (aka “end gaps”).
Compare DNASTAR Lasergene Packages
| MOST POPULARDNASTAR Lasergene | ||||
| Lasergene Molecular Biology | Lasergene Genomics | Lasergene Protein | ||
|---|---|---|---|---|
| Included Applications | ||||
| SeqBuilder Pro | ||||
| SeqMan Ultra | ||||
| MegAlign Pro | ||||
| GeneQuest | ||||
| GenVision | ||||
| SeqNinja | ||||
| SeqMan NGen | ||||
| ArrayStar | ||||
| GenVision Pro | ||||
| Protean 3D (+1 prediction per Nova Application) |
||||
| DNASTAR Navigator | ||||
| Supported Workflows | ||||
| Integrates with | ||||
| See Pricing | See Pricing | See Pricing | See Pricing | |