If you are working with sequence data, you may export regions of the template sequence(s) corresponding to currently selected genes, peaks or fragments by selecting Data > Export Sequences. Reads exported via this command are intended for further analysis using applications such as SeqMan NGen, which may have different alignment criteria from ArrayStar. As such, ArrayStar will export all reads which appear to be usable by assembly or other sequence analysis methods, based on these reads having partial similarity to the target sequences (i.e. genes). This is intended to be a permissive export method and may export reads which did not meet the threshold for quantification within ArrayStar.
Note: To instead export the exact matches used during ArrayStar quantification, check the Alignment Files box in the Export tab of QSeq Advanced Options.
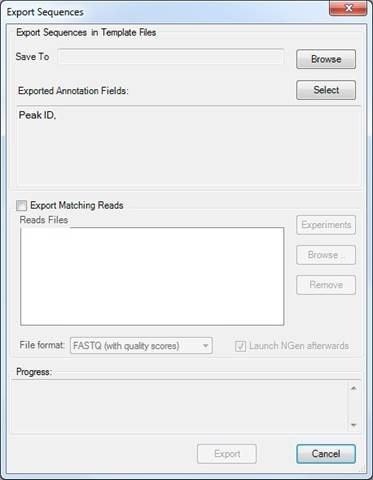
After choosing Data > Export Sequences, the following dialog will appear:

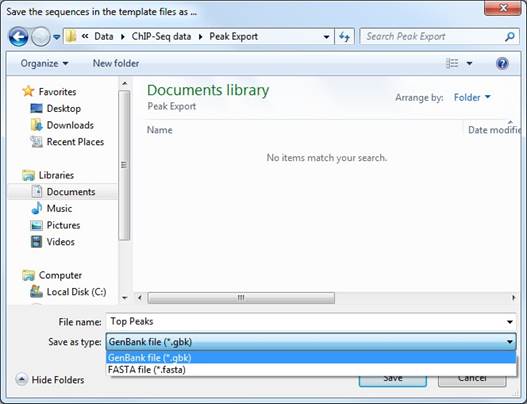
Click Browse to open the following dialog. From here, navigate to the location where you would like the exported file to be saved, choose whether to save a GenBank (.gbk) or FASTA (.fasta) file, and name the file. Click Save to apply the changes for the output file and return to the Export Sequences dialog or Cancel to close this dialog without making any changes.
ArrayStar will export a single GenBank or FASTA file which contains all the regions of the template sequence(s) that correspond to the selected genes, peaks, or fragments as individual sequences.
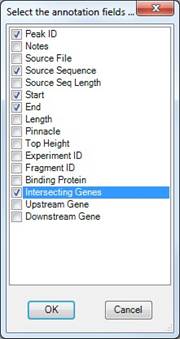
Click Select to open the following dialog. Here you can select annotations to be included in the export by check the boxes next to the annotation names. Click OK to apply the changes or click Cancel to close this dialog without making any changes.
You may also elect to export the reads that map to the selected regions of the template by checking the box next to Export Matching Reads.
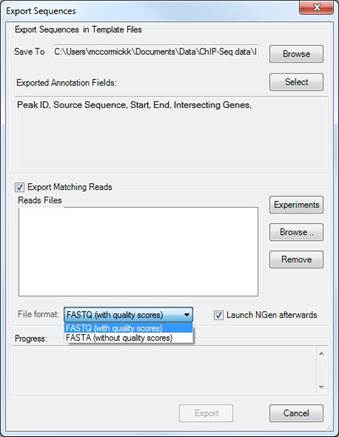
Click Experiments to choose the experiment(s) whose reads you wish to export. By default, reads are exported for all experiments.
Click Browse to navigate to the sequence files that contain the sequence data. Once you have selected sequence files for the export, they will be added to the Reads Files list. You may remove files from this list by highlighting them and selecting Remove.
If you are exporting matching reads, you may specify whether to export them with or without quality scores.
Once you are satisfied with your export settings, click Export to begin the export process. Click Cancel to exit this dialog without exporting sequences.