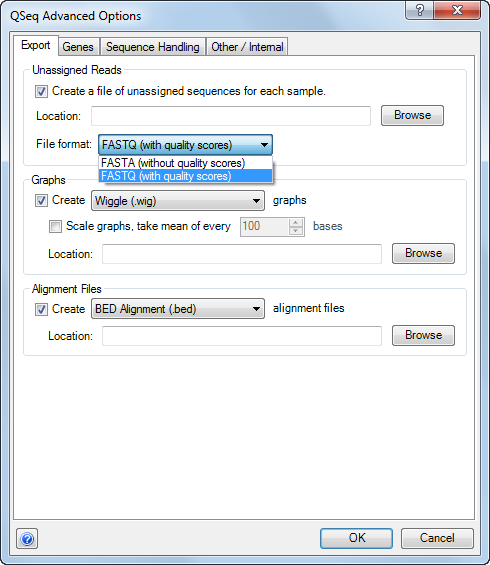

•Check the box under Unaligned Reads to create a FASTA file containing the reads that were not assigned to a template during the processing. Use the Location field to specify the directory where the file will be saved. You may choose to save in FASTA or FASTQ File format, depending upon whether or not you want to export quality scores for the unassigned reads.
•Check the box in the Graphs section to export GenVisionTM maps or Wiggle track files that show the reads mapped at each position along the template(s) as peaks. One graph will be exported per template sequence. To limit the size of these files, you may wish to Scale graphs. If this option is checked, QSeq will take the mean of the number of reads mapped to the template over the specified number of bases. Use the Location field to specify the directory where the files will be saved.
GenVision maps can be viewed and edited in DNASTAR’s GenVision. One .gnv GenVision file and one .txt data file will be exported per template sequence. Wiggle graphs can be loaded as custom tracks and viewed in the UCSC Genome Browser. One .wig file will be exported per template sequence.
Note: If you are interested in trying or purchasing GenVision, please visit www.dnastar.com or contact your Sales Representative.
•Check the box and use the drop-down menu in the Alignment Files section to create GFF or BED alignment files that contain the results of QSeq’s mapping. One file will be exported per experiment. Use the Location field to specify the directory where the file should be saved.
Note that the exported graph and alignment files do not contain repeated reads. This may cause a discrepancy between the exported file and the QSeq results in the Gene Table, Peak Table and Fragment Table if repeat handling is used during preprocessing. For more information on repeat handling in QSeq, see the advanced options for Internal and Peak Detection settings.