To export a set from the Set List and save it as a text (.txt) or Variant Call Format (.vcf) file:
1) Create and name a set of genes, SNPs, exons, etc.
Note: To export a Variant Call Format (VCF) file using this method, you must create a SNP set from within a Variants project.
2) Open the Set List view by selecting Data > Show Set List.

3) Select one of the sets. Then, do any of the following:
•Right-click and choose Export.
•Select File > Export Selected Set.
•Click the Export Selected Set tool .
.
4) In the Export Names from Set dialog, specify a location, name and file type.
•If you selected a gene set, the names for each gene in the set will be exported as a .txt file. You can assign the data column to be used as the gene name by modifying Gene Information Settings.
•If you selected a SNP set, all SNPs in the set will be exported as a Variant Call Format (.vcf) file.
5) Press the Save button. Unless a multiple-assembly project is open, this concludes the export procedure.
6) (Multiple-assembly project only). If you are working with a project containing both RNA-Seq and variant data, the Choose experiments dialog will open.
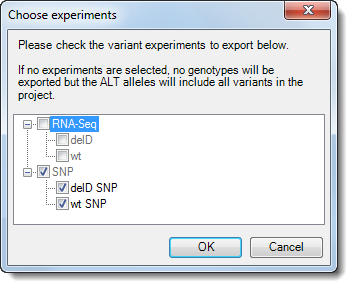
7) Check or uncheck experiment boxes, as desired. If one or more boxes are checked, ArrayStar will export a sample VCF file for each of the checked items. If no boxes are checked, ArrayStar will export an annotation-only VCF of the type exported in Lasergene version 13.
See VCF File Columns for a description of three columns that make up part of an exported .vcf file.
Note: For an alternative way to export a VCF file in ArrayStar, see Exporting Table Rows as Text CSV or VCF Files.