To export selected rows from an ArrayStar table:
1) Add or remove column types and arrange the order of columns as desired.
2) Select the rows you wish to export so they are highlighted in blue. Use Shift+click to select consecutive rows; Ctrl/Cmd+click to select non-consecutive rows; or Ctrl/Cmd+click to select all rows.
3) Right-click on any highlighted row and choose Export Selected Table Rows. Alternatively, choose File > Export Selected Table Rows from the main menu.
4) In the Export Names from Set dialog, specify a location, name and file type. Save as type options always include tab-delimited (.txt) and comma-delimited (.csv) text. Regardless of the table from which you are exporting, these choices will export the actual contents of the table rows. If you are exporting from the SNP Table, you may also choose to export the selected data as a Variant Call Format (.vcf) file.
5) Press the Save button. Unless a multiple-assembly project is open, this concludes the export procedure.
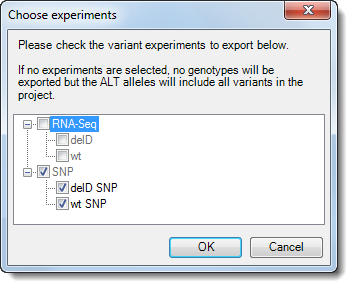
6) If you are exporting a VCF from the SNP Table and working with a multiple-assembly project containing both RNA-Seq and variant data, the Choose Experiments dialog will open.

7) Check or uncheck experiment boxes, as desired. If one or more boxes are checked, ArrayStar will export a sample VCF file for each of the checked items. If no boxes are checked, ArrayStar will export an annotation-only VCF of the type exported in Lasergene version 13.
See VCF File Columns for a description of three columns that make up part of an exported .vcf file.
Note: For an alternative way to export a VCF file in ArrayStar, see Exporting Sets as Text or VCF Files.