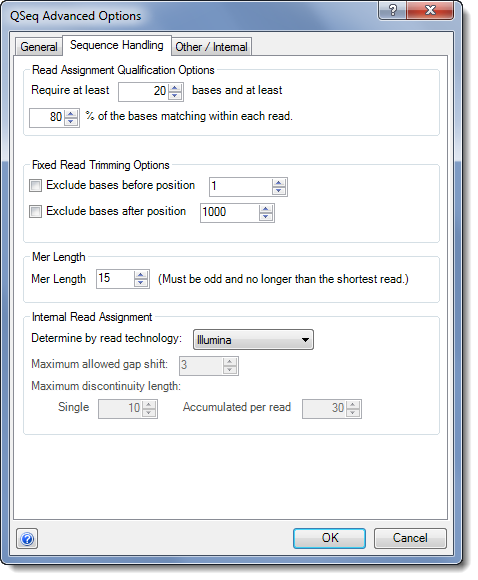

•The settings under Read Assignment Qualification Options allow you to specify the minimum requirements necessary for a read to be considered as a match for a template sequence. Here you can:
o Change the minimum number of bases that must match the template within each read.
o Change the minimum match percentage to specify what percentage of the read must match the template.
Reads will only be considered if both the total number of bases and percentage of bases within each read meet these thresholds. In addition to meeting these requirements, a read must not be considered a repeat and must contain matching mers of the correct length in order to be mapped to the template sequence.
Note: Read Assignment Qualification uses either the read length or the target (i.e., "exon") length, whichever is shorter.
•In the section Fixed Read Trimming Options, you may choose to trim in one or both of the following ways:
o Check the box next to Exclude bases before position to exclude the first n number of bases from the beginning of each read.
o Check the box next to Exclude bases after position to exclude the last n bases from the end of each read.
•Specify the Mer Length that you would like ArrayStar to use for matching your reads to the template sequences. This value must be odd and no longer than the shortest read in your data set.
Note: Reads with mers that match to the template may or may not be assigned as matches, depending upon the settings in this dialog and in repeat filtering.
•Select the desired options from the Internal Read Assignment panel:
•Choose the appropriate sequencing technology from the Determine by read technology drop-down menu.
o Maximum allowed gap shift – The maximum number of gaps allowed in a read for that read to be mapped.
o Maximum discontinuity length - Single = the maximum number of mismatching bases in any single stretch of bases for that read to be mapped. Accumulated per read = the maximum number of mismatching bases across the entire read for that read to be mapped.