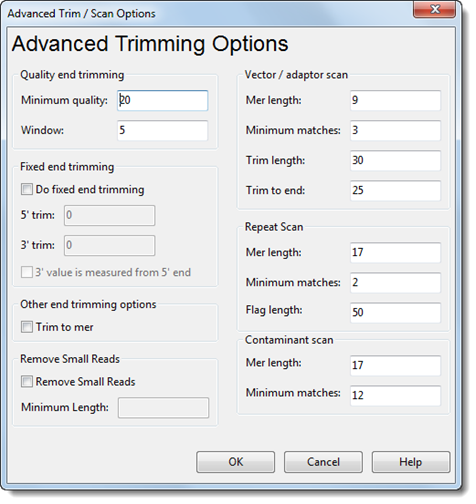
From the Read Options dialog, clicking the Advanced Trim/Scan Options button brings you to the Advanced Trim/Scan Options dialog. This dialog allows you to view and modify trimming parameters and vector, repeat and contaminant scanning parameters.

Quality End Trimming Settings:
•Minimum quality – The minimum averaged quality score of the evaluated window that is required in order to be considered low-quality.
•Window – The length of the window to be used for averaging quality scores.
Fixed End Trimming Settings:
•Do fixed end trimming – Check this box to implement pre-assembly fixed end data trimming. Enter the number of base pairs you wish to trim in the 5’ trim and 3’ trim fields.
The values entered for 5’ trim and 3’ trim are used differently, depending on whether 3’ value is measured from 5’ end is selected. If it is not selected, then the 5’ trim and 3’ trim values will indicate the number of bases for SeqMan NGen to trim from the respective ends of each read. If it is selected, then the 5’ trim and 3’ trim values will indicate the specific coordinates to which reads should be trimmed.
Other End Trimming Options:
•Trim to mer – Check this box to trim the reads to the matching mer within the read. For each read, SeqMan NGen looks for mers that exist in the template (for templated assemblies) or in any other read in the assembly (for de novo assemblies). It then sets the trimming for the read to the start of the first mer found and the end of the last mer found. Trimming to mer may be useful when assembling data without accurate quality scores or data with very short linkers.
Remove Small Reads:
•Remove small reads – Check this box to remove reads that do not meet a threshold that you specify. If you check this box, you must enter a Minimum length in the box provided.
Vector/Adapter Scan Settings:
•Mer length – The minimum length of a mer required to be considered an exact match when searching for vector.
•Minimum matches – The minimum number of matching mers required to start an alignment.
•Trim length – The minimum length required for a mer to be considered as a match for vector trimming.
•Trim to end – The distance to the endpoint where trimming will go all the way to the end of the sequence.
Repeat Scan Settings:
•Mer length – The minimum length of a mer required to be considered an exact match when scanning for repeats.
•Minimum matches – The minimum number of matching mers required to be considered a repeat.
•Flag length – The minimum length required for a mer to be flagged as a repeat.
Contaminant Scan Settings:
•Mer length – The minimum length of a mer required to be considered an exact match when scanning for contaminants.
•Minimum matches – The minimum number of matching mers required to mark the sequence as a contaminant.
Once you are finished, click OK to save your changes and return to the Read Options dialog, or Cancel to discard changes before returning.