 .
. The Info Pane appears to the right of the Heat Map, and gives information about the data displayed.
The Info Pane is separated from the rest of the Heat Map view by a splitter bar
which can be dragged to resize the window. The Info Pane may be hidden or shown
by using the Hide Info Pane tool  .
.
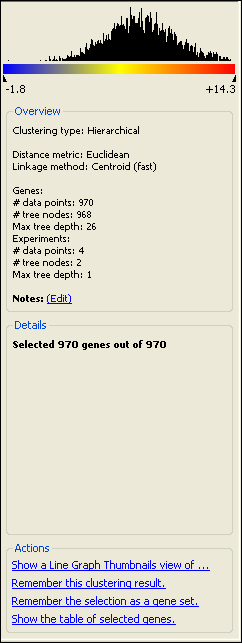
The color scale:
The scale at the top of the Info Pane is used to display the signals (e.g., expression levels) in the current Heat Map view is displayed at the top of the Info Pane.
A histogram of the data distribution in the Heat Map is displayed above the color scale. As selections are made on the Heat Map, the selected genes are shown in gray within the histogram.
If outlier cutoff values have been assigned, the cutoff regions will be displayed in both the histogram and the color scale in blue and red, by default.
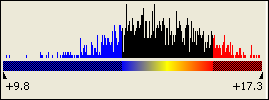
The minimum and maximum signal values are shown beneath the color scale.
The color scale can be displayed as an absolute or relative
scale by clicking on the Heatmap Color Scale tool  and selecting All Project Data
(absolute scale), or Current Heat Map Data Points (relative
scale).
and selecting All Project Data
(absolute scale), or Current Heat Map Data Points (relative
scale).
The relative color scale uses only the range of signal values from the data currently displayed in the Heat Map:
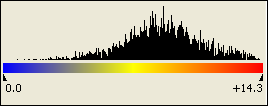
The absolute color scale uses the range of signal values from your entire project.
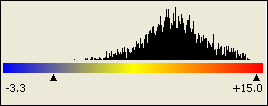
If the absolute scale is used, two triangular marks below the color scale mark the minimum and maximum expression values in the current view, as shown below:
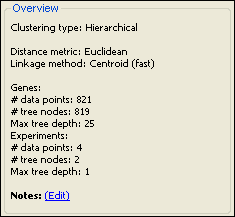
The Overview section:
This section displays the Clustering Type that was used for the current data set, as well as the distance metric and linkage method, if applicable. Also shown is the number of data points plotted in the Heat Map, the number of tree nodes, and the maximum tree depth.
You may add notes for your clustering by clicking on the “Notes: (Edit)” link.
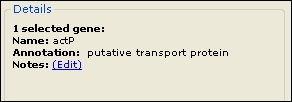
This section displays information about the genes currently selected in the Heat Map.
If no genes are selected, the number of genes in the clustering is displayed.

If you have selected exactly one gene, you can add a note about it by clicking on the “Notes: (Edit)” hyperlink.
When one gene is selected, you can also right-click within the Details panel to access the following context menu commands:
|
Annotation requirements |
Right-click command |
Command description |
|
No annotations necessary |
Modify gene information settings |
Shortcut to Data > Show Gene Table. |
|
Gene must contain annotations |
Quick search for |
Finds other genes in your project with the same annotation. |
|
Advanced search for |
Launches the Advanced Filtering dialog with the specified annotation already populated as a search criterion. |
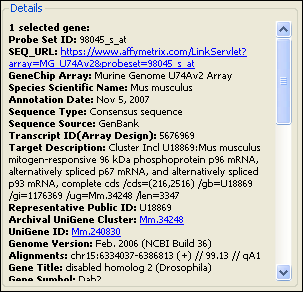
Some annotations for the selected gene may have special features:
Hover your mouse over annotations shown in color to display additional information.
Click on hot links (shown in color with an underline) to link to additional information for that annotation online.
For Gene Ontology terms, hovering your mouse over a term will display a more detailed description of the term. Clicking on a Gene Ontology term will link you to the full description online.
If multiple genes are selected, then the number of genes selected is shown in this section.
In addition, as your cursor moves over the Heat Map, details for each gene are displayed as you mouse-over them, including the gene name, annotations, notes, and the signal for the experiment your mouse is hovering over.
The Actions section:
The Actions section is located at the bottom of the Info Pane and provides hyperlinked shortcuts for various tasks. Options vary depending on the view and workflow. See Actions Section Hyperlinks for a description of each hyperlink.
Note: If the Actions section is not visible, you may first need to select data from the left side of the view.