The QSeq Peak Finder is based on the ERANGE 3.1 Algorithm for ChIP-Seq and RNA-Seq Analysis (see Mortazavi et al., 2008). This peak detection algorithm calculates peaks in a normalized reads-per-million space. Features of this algorithm include simple read shifting and repeat read handling. This algorithm also considers the directionality of reads when calling peaks.
The ChIP-Seq Peak Finder reports the number of reads within the peak as the score for that peak. In addition, the ChIP-Seq Peak Finder reports a P-value that contains the likelihood of a given region being a "true" peak. The P-value is based on how many peaks QSeq would have called in the control lane.

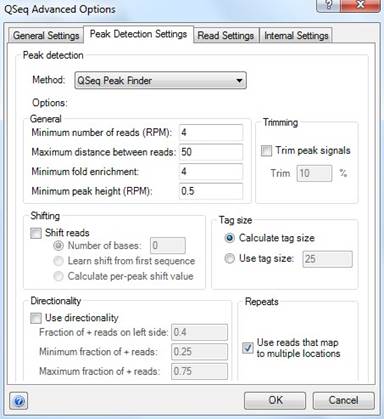
If you choose QSeq Peak Finder as the peak detection method, the following options will be available:
•The General settings specify the requirements a region must satisfy to be considered a peak. A region must contain a Minimum number of reads and have a Minimum peak height to be considered a peak. Both of these values are specified in "Reads Per Million.” So, for example, if a lane contains 6 million reads and the Minimum number of reads value is set to the default of 4, a region would need to contain a minimum of 24 reads to be considered as a peak. The Maximum distance between reads value specifies how close a read must be to the nearest read in a region to be considered part of that region. The Minimum fold enrichment value specifies how many more reads must be in a region versus the same region in the control lane to be considered a peak.
Note: Control data can only be used for ChIP-Seq experiments. You may specify control experiments in the Create Binding Proteins dialog of the Project Setup Wizard.
•If Trimming is used, QSeq will adjust the start and end positions of each peak so that the height, or depth of reads, in those positions is equal to or greater than the specified percentage of the maximum height of the peak.
•The Shifting settings allow QSeq to compensate for the tag shift artifact in ChIP-Seq data. If Shift Reads is checked, you have the option to specify a Number of bases to shift the reads. Alternatively, you can specify that QSeq calculate the shift, either based on the first template sequence by checking Learn shift from first sequence, or on a peak by peak basis by checking Calculate per-peak shift value.
•The Tag Size settings define the length of read used for peak detection. The ChIP-Seq Peak Finder only uses the start position of each read and treats all reads as though they were the same length.
•The Directionality settings allow you to define minimum requirements for the proportions of forward and reverse reads within each peak. If Use directionality is checked, any peaks not meeting these criteria will be discarded.
•In the Repeats section, check Use reads that map to multiple locations if you want QSeq to consider reads that map to multiple locations during peak detection. If repeat reads are considered, you will be able to see the proportion of the peak score that comes from the repeated reads in the raw_repeat_count signal column of the Peak Table after processing the data. You may uncheck this option if you only want QSeq to consider reads that map to unique locations.