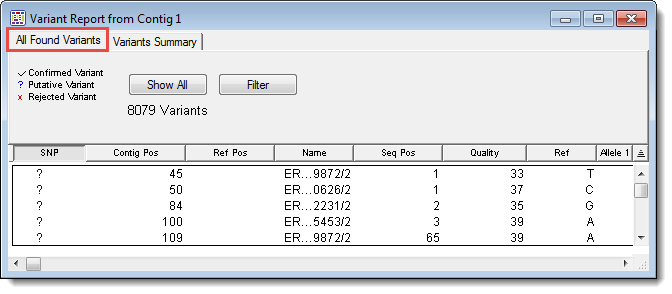
To access the All Found Variants Report for a selected contig, use Variant > Variant Report, and then click on the All Found Variants tab. If no contig is currently selected, you must first select one from the Project Summary window before opening the report.
Note: The All Found Variants Report tab is disabled for BAM-based projects. For these projects, only the Variants Summary tab is active.

For information about items in the table header, see Working with Variant Reports.
Each row in the All Found Variants report lists information for a single variant base in a sample sequence. The columns for each row include:
|
Column Name/Icon |
Description |
|
SNP |
A question mark indicates that all variant bases in the aligned column are putative, a checkmark that all are confirmed, an ‘X’ indicates that all in the column are rejected as actual variants, and a dash is displayed if the column contains some combination of putative, confirmed, and rejected variants. Click on a mark to change the evaluation for all variants in the aligned column. |
|
Contig Pos |
The position of the variant in the contig including gaps. |
|
Ref Pos |
The position of the variant in the reference sequence excluding gaps. |
|
Name |
The name of the sample sequence that contains the variant. |
|
Seq Pos |
The position of the variant in the sample sequence. |
|
Quality |
The quality score of the variant base. For trace data, including gaps, Q/n scores will be displayed. If imported quality scores are available, they will be displayed. (For non-trace data, gaps will show a quality score equal to the average quality score of its two adjacent bases.) |
|
Ref |
The reference sequence base in this position. If there is no reference sequence present, the Ref column will display the most frequently occurring non-ambiguous base at this position. If no such base exists, the consensus base at this position will be shown. |
|
Allele 1-4 |
The variant bases in these positions. |
To display all variants and negate any previous filtering (see below), click the Show All button.
To display only those variants of most interest to you, click the Filter button. See Filtering Variants in the Reports for detailed information.
To navigate between variants in the sequences, open the Alignment View (Contig > Alignment View) and arrange the windows so you can see the Alignment View and the Variant Report simultaneously. Select a row in the Variant Report, then click on the up or down arrow key on your keyboard to move to the previous or next variant in both the Variant Report and the Alignment View.
To locate the variant in the Alignment View, double click on a row in the Variant Report. The Alignment View will open at the location of the selected variant base.