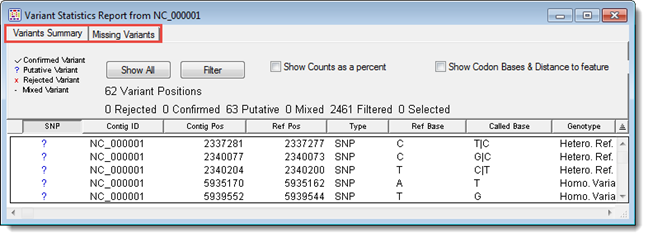
SeqMan Pro’s variant reports contain information about the variants in one or more contigs.
To access the variant reports for the selected contig(s), choose Variant > Variant Report. This command is accessible from the Alignment View, Strategy View or after selecting a contig, scaffold or group of Unlocated contigs in the Project Summary window. If a scaffold or group of Unlocated contigs is selected, SeqMan Pro scans all the contigs included in the selection.
If you are working with a BAM-based project for which variants have not been pre-calculated, SeqMan Pro will not calculate them. Instead, SeqMan Pro returns an error message informing you that you can only calculate variants for these projects during assembly (e.g. with SeqMan NGen).
Note: You can also use SeqMan NGen’s “Import BAM file” option to calculate variants for one or more existing BAM files without having to do a full assembly.
The ensuing Variant Report window contains one or two tabs, depending on the project type. The All Found Variants is available for non-BAM-based projects only, while the Variants Summary is available for all project types. See those topics for detailed information about each column in the report.
Table headers for different tabs may list one or more of the following counts.
|
Name |
Description |
|
SNP Positions |
Total collapsed number of variant positions (i.e., the number of reference positions with a called variant in one or more samples). |
|
Rejected |
Number of variants with ‘x’ in first column. |
|
Confirmed |
Number of variants with checkmark (P) in first column. |
|
Putative |
Number of variants with question mark (?) in first column. |
|
Mixed |
Number of variants marked as a mixture of putative, confirmed and/or rejected. |
|
Filtered |
Number of variants currently filtered out of the report using settings accessed via the Filter button. |
|
Selected |
Number of variants currently selected in the Report.
To see the total number of “uncollapsed” variant positions (i.e., the number of variant positions plus the number of samples with a variant at each position), select all entries in the Variant Report using Ctrl/Cmd+A. Then observe the number Selected. If you are working with multi-sample data, you can see all the variants for a particular MID sample by sorting by MID column and selecting only the rows for the MID sample of interest. |
By default, the information displayed in the reports uses the filters selected in SeqMan NGen prior to assembly. These filters can be changed in SeqMan Pro using the Filter button (see Filtering Variants in the Reports).

Depending on the project type, one or two checkboxes will appear in the Variants Summary report header. The Show Counts as a percent checkbox toggles between showing number (Cnt) or percentage (%)in the base columns. The Show Codon Bases checkbox (only available for BAM-based projects) toggles between showing the Codon / Coding Feature Distance and the DNA Change / Amino Acid Change columns.
In the body of the report, columns can be displayed, hidden or rearranged, as desired. See Sorting and Rearranging Variant Report Columns for details.
To update the Variant Report, select the menu command Variant > Update Variant Report. This command is disabled unless an .sqd file is open and is normally hidden from view in the Variant menu. To reveal the command, hold down the Alt (Win) or Option (Mac) key while clicking on the Variant menu.
Right-click in either Variant Report to access context menu shortcuts to main menu commands:
|
Command |
Shortcut to: |
|
Copy |
Edit > Copy |
|
Confirm Variant |
Variant > Confirm Variant |
|
Reject Variant |
Variant > Reject Variant |
|
Putative Variant |
Variant > Putative Variant |
|
Select All of Type |
Variant > Select All of Type |
|
Show/Hide Column |
View > Show/Hide Column |
|
Sort |
View > Sort |